1KM0
 
 | |
4L76
 
 | | Ca2+-bound E212Q mutant MthK RCK domain | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK, SODIUM ION | | Authors: | Smith, F.J, Rothberg, B.S. | | Deposit date: | 2013-06-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Structural basis of allosteric interactions among Ca(2+)-binding sites in a K(+) channel RCK domain.
Nat Commun, 4, 2013
|
|
4LC8
 
 | | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4L75
 
 | |
4NX5
 
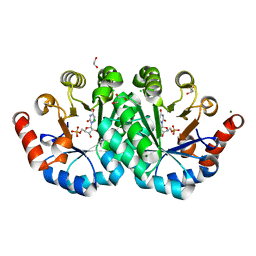 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-AZA URIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate
To be Published
|
|
4O11
 
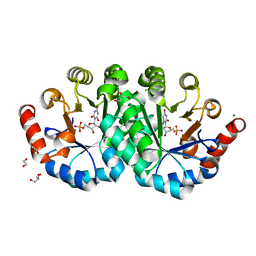 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate
To be Published
|
|
4NT0
 
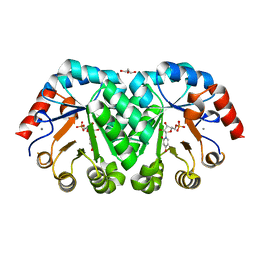 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate | | Descriptor: | 4-hydroxy-1-(5-O-phosphono-beta-D-ribofuranosyl)pyridin-2(1H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate
To be Published
|
|
4EI2
 
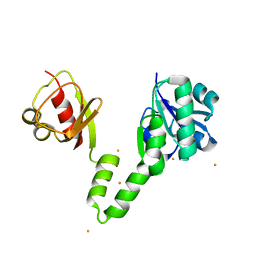 | |
1KLZ
 
 | |
1KM1
 
 | |
1KM5
 
 | |
4HYO
 
 | | Crystal Structure of MthK Pore | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4O8R
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate
To be Published
|
|
4FZP
 
 | |
4FZO
 
 | |
3SY5
 
 | | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor 6azaUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-07-15 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Crystal structure of the mutant S127A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor 6azaUMP
To be Published
|
|
6OLY
 
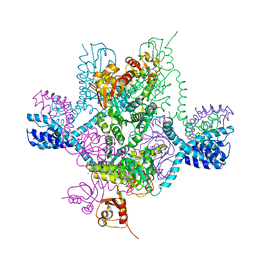 | | Full-length MthK channel at 3.1 angstrom resolution | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK | | Authors: | Rothberg, B.S. | | Deposit date: | 2019-04-17 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Molecular mechanism of a potassium channel gating through activation gate-selectivity filter coupling.
Nat Commun, 10, 2019
|
|
6Y1X
 
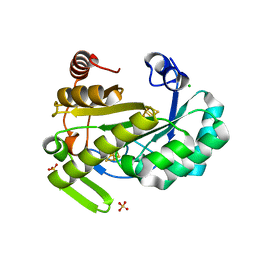 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
6PSP
 
 | |
8QUU
 
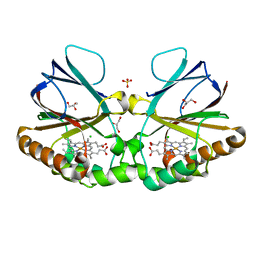 | | Crystal structure of chlorite dismutase at 3000 eV based on spherical harmonics absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QVB
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on a combination of spherical harmonics and analytical absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUV
 
 | | Crystal structure of chlorite dismutase at 3000 eV with no absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUZ
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on analytical absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
6QP1
 
 | | Crystal structure of the PLP-bound C-S lyase in the external aldimine form from Staphylococcus hominis complexed with an inhibitor, L-cycloserine. | | Descriptor: | Aminotransferase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
6QP2
 
 | | Crystal structure of the PLP-bound C-S lyase from Staphylococcus hominis | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE, Polyhistidine tag | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
