4GTC
 
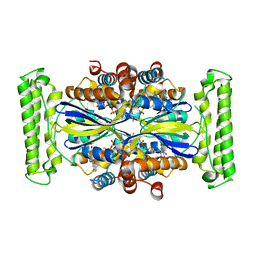 | | T. Maritima FDTS (E144R mutant) plus FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A, Prabhakar, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TOB
 
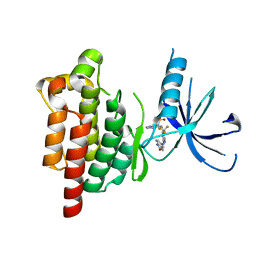 | |
4GT9
 
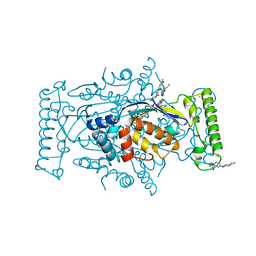 | | T. Maritima FDTS with FAD, dUMP and Folate. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTL
 
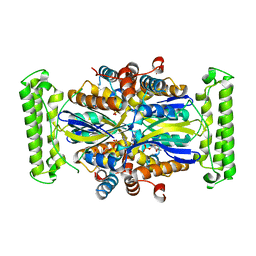 | |
4ERP
 
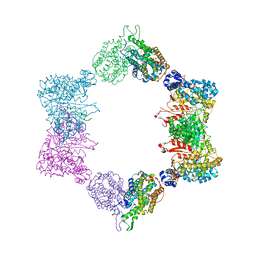 | |
5TO8
 
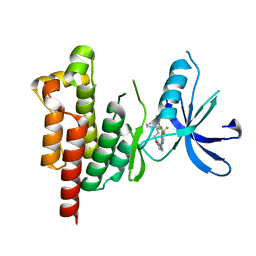 | | Selectivity switch between FAK and Pyk2: Macrocyclization of FAK inhibitors improves Pyk2 potency | | Descriptor: | 25-(methylsulfonyl)-8-(trifluoromethyl)-5,17,18,21,22,23,24,25-octahydro-12H-7,11-(azeno)-16,13-(metheno)pyrido[3,2-i]pyrrolo[1,2-q][1,3,7,11,17]pentaazacyclohenicosin-20(6H)-one, Protein-tyrosine kinase 2-beta | | Authors: | Newby, Z.E. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9849 Å) | | Cite: | Selectivity switch between FAK and Pyk2: Macrocyclization of FAK inhibitors improves Pyk2 potency.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
4O0A
 
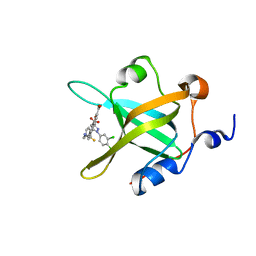 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)-2-chlorophenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J. Med. Chem., 56, 2013
|
|
3HKY
 
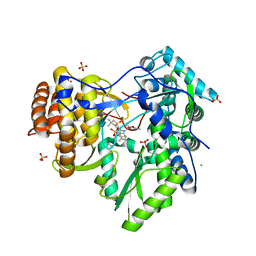 | | HCV NS5B polymerase genotype 1b in complex with 1,5 benzodiazepine 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
4I5Y
 
 | |
4OOP
 
 | | Arabidopsis thaliana dUTPase with with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
4CSG
 
 | |
3PO8
 
 | |
6MIS
 
 | | Native ananain in complex with E-64 | | Descriptor: | Ananain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Yongqing, T, Wilmann, P.G, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Determination of the crystal structure and substrate specificity of ananain.
Biochimie, 166, 2019
|
|
6IZP
 
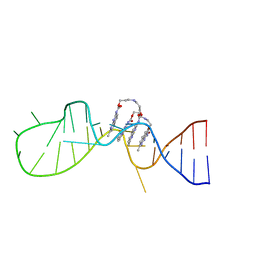 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
4FRV
 
 | | Crystal structure of mutated cyclophilin B that causes hyperelastosis cutis in the American Quarter Horse | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
4FHA
 
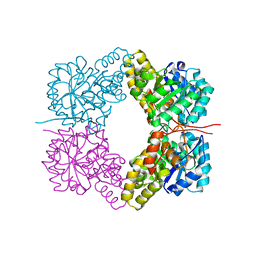 | | Structure of Dihydrodipicolinate Synthase from Streptococcus pneumoniae,bound to pyruvate and lysine | | Descriptor: | Dihydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Perugini, M.A, Dogovski, C, Parker, M.W, Gorman, M.A. | | Deposit date: | 2012-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure, Function, Stability and Knockout Phenotype of Dihydrodipicolinate Synthase from Streptococcus pneumoniae
To be Published
|
|
4FRU
 
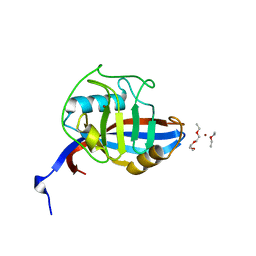 | | Crystal structure of horse wild-type cyclophilin B | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
6MOY
 
 | |
4E6W
 
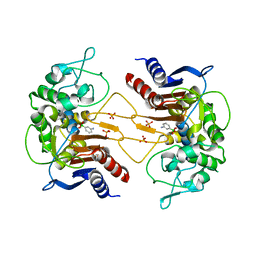 | | ClbP in complex with 3-aminophenyl boronic acid | | Descriptor: | ClbP peptidase, M-AMINOPHENYLBORONIC ACID, PHOSPHATE ION | | Authors: | Cougnoux, A, Delmas, J, Bonnet, R. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The NRP peptidase ClbP as a target for the inhibition
of genotoxicity, cell proliferation and tumorogenesis
mediated by pks-harboring bacteria
To be Published
|
|
4OOQ
 
 | | apo-dUTPase from Arabidopsis thaliana | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, SULFATE ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
4G75
 
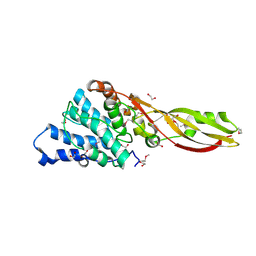 | | Structure of PaeM, a colicin M-like bacteriocin produced by Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, phosphodiesterase | | Authors: | Touze, T, Graille, M, Mengin-Lecreulx, D. | | Deposit date: | 2012-07-20 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of PaeM, a Colicin M-like Bacteriocin Produced by Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
4GGJ
 
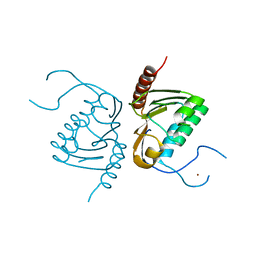 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) | | Descriptor: | Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
3SR6
 
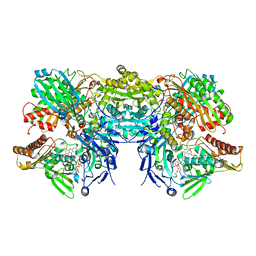 | | Crystal Structure of Reduced Bovine Xanthine Oxidase in Complex with Arsenite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Crystal Structure of Arsenite-Inhibited Xanthine Oxidase: Mu-Sulfido,Mu-Oxo Double Bridge between Molybdenum and Arsenic in the Active Site.
J.Am.Chem.Soc., 133, 2011
|
|
1D5Z
 
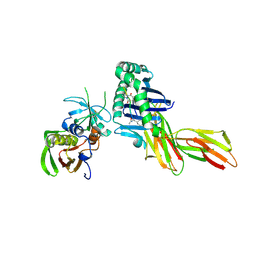 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | PROTEIN (ENTEROTOXIN TYPE B), PROTEIN (HLA CLASS II HISTOCOMPATIBILITY ANTIGEN), PROTEIN (PEPTIDOMIMETIC INHIBITOR) | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1D6E
 
 | | CRYSTAL STRUCTURE OF HLA-DR4 COMPLEX WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, PEPTIDOMIMETIC INHIBITOR | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-13 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
