8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8SF8
 
 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
6TSP
 
 | | Marasmius oreades agglutinin (MOA) inhibited by zinc | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: a protease caught in flagranti
Curr Res Struct Biol, 2, 2020
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UMH
 
 | | Energetic robustness to large scale structural dynamics in a photosynthetic supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Harris, D, Toporik, H, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex.
Nat Commun, 14, 2023
|
|
6HMG
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
7UVD
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Sco GlgEI-V279S in complex with cyclohexyl carbasugar
To Be Published
|
|
6BHQ
 
 | | Mouse Immunoglobulin G 2c Fc fragment with complex-type glycan | | Descriptor: | GLYCEROL, Igh protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Falconer, D, Barb, A. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1.
PLoS ONE, 13, 2018
|
|
6TOZ
 
 | | Crystal structure of Bacillus paralicheniformis alpha-amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETIC ACID, Amylase, ... | | Authors: | Rozeboom, H.J, Janssen, D.B. | | Deposit date: | 2019-12-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the starch surface binding site on Bacillus paralicheniformis alpha-amylase.
Int.J.Biol.Macromol., 165, 2020
|
|
4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
5JCQ
 
 | |
3J9F
 
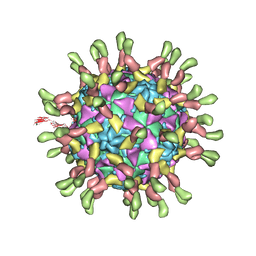 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
4WMT
 
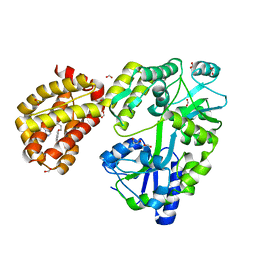 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
5NGE
 
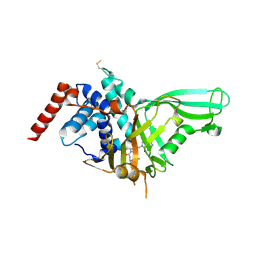 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
8DDF
 
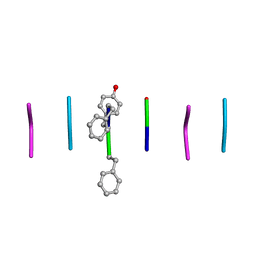 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
5EIV
 
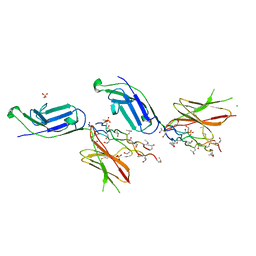 | | Crystal structure of complex of osteoclast-associated immunoglobulin-like receptor (OSCAR) and a synthetic collagen consensus peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhou, L, Blaszczyk, M, Chirgadze, D, Bihan, D, Farndale, R.W. | | Deposit date: | 2015-10-30 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Structural basis for collagen recognition by the immune receptor OSCAR.
Blood, 127, 2016
|
|
6F92
 
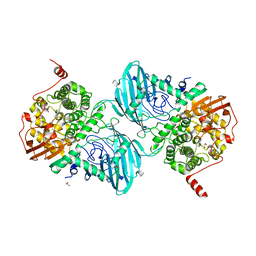 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7JW0
 
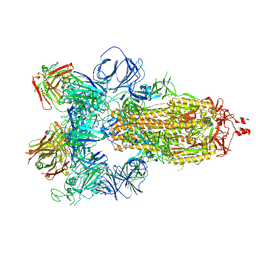 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7K43
 
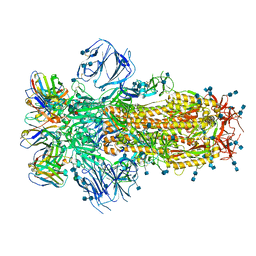 | |
7MQO
 
 | | The insulin receptor ectodomain in complex with a venom hybrid insulin analog - "head" region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, ... | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
4ZAK
 
 | | Crystal structure of the mCD1d/DB06-1/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Birkholz, A.M. | | Deposit date: | 2015-04-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
J Immunol., 195, 2015
|
|
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
8OVV
 
 | |
