8DA8
 
 | | Coevolved affibody-Z domain pair LL2.c1 | | Descriptor: | Affibody LL2.FIIK, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA7
 
 | | Coevolved affibody-Z domain pair LL1.c6 | | Descriptor: | Immunoglobulin G-binding protein A, MALONATE ION, affibody LL1.FIFV | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAC
 
 | | Coevolved affibody-Z domain pair LL2.c22 | | Descriptor: | Affibody LL2.FILV, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAA
 
 | | Coevolved affibody-Z domain pair LL2.c7 | | Descriptor: | Affibody LL2.FIVK, Immunoglobulin G-binding protein A, MALONATE ION | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAB
 
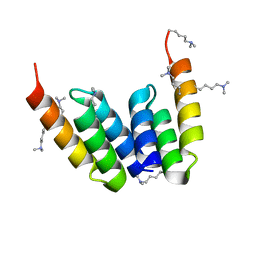 | | Coevolved affibody-Z domain pair LL2.c17 | | Descriptor: | Affibody LL2.IVVY, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.134 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
5F3W
 
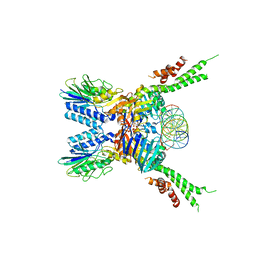 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
6UB3
 
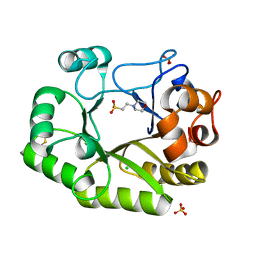 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) with laminaribiose at the surface-binding site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
5C1B
 
 | |
6UAV
 
 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SULFATE ION | | Authors: | Santos, C.R, Costa, P.A.C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB1
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites | | Descriptor: | GLYCOSIDE HYDROLASE, beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
7FEH
 
 | | Crystal structure of human DDR1 in complex with CH5541127 | | Descriptor: | Epithelial discoidin domain-containing receptor 1, N-[(5-chloranyl-2-ethylsulfonyl-phenyl)methyl]-3-piperazin-1-yl-5-(trifluoromethyloxy)benzamide, NITRATE ION | | Authors: | Fukami, T.A, Kadono, S, Matsuura, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Novel potent and highly selective DDR1 inhibitors from integrated lead finding
Medicinal Chemistry Research, 32, 2023
|
|
4QIA
 
 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
6UAR
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAU
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glyco_hydro_cc domain-containing protein, ZINC ION, ... | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB0
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UBB
 
 | | Crystal structure of a GH128 (subgroup VI) exo-beta-1,3-glucanase from Aureobasidium namibiae (AnGH128_VI) with laminaribiose at the surface-binding site | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Vieira, P.S, Domingues, M.N, Cordeiro, R.L, Tomazini, A, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
7L7K
 
 | |
6UB7
 
 | | Crystal structure of a GH128 (subgroup V) exo-beta-1,3-glucanase from Cryptococcus neoformans (CnGH128_V) | | Descriptor: | Glyco_hydro_cc domain-containing protein, POTASSIUM ION | | Authors: | Santos, C.R, Costa, P.A.C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAT
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose | | Descriptor: | Glyco_hydro_cc domain-containing protein, ZINC ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAY
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) | | Descriptor: | GLYCOSIDE HYDROLASE | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB8
 
 | | Crystal structure of a GH128 (subgroup VI) exo-beta-1,3-glucanase from Aureobasidium namibiae (AnGH128_VI) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein | | Authors: | Santos, C.R, Vieira, P.S, Domingues, M.N, Cordeiro, R.L, Tomazini, A, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
5GGU
 
 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGS
 
 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
6UAQ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SODIUM ION | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAW
 
 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, SULFATE ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Costa, P.A.C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
