2FKG
 
 | | The Crystal Structure of Engineered OspA | | Descriptor: | Outer Surface Protein A | | Authors: | Makabe, K, Terechko, V, Gawlak, G, Yan, S, Koide, S. | | Deposit date: | 2006-01-04 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F37
 
 | |
4RPD
 
 | |
4ROX
 
 | |
2CEH
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
2CM2
 
 | | Structure of Protein Tyrosine Phosphatase 1B (P212121) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
3ZV8
 
 | |
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | Descriptor: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZXR
 
 | |
2C8O
 
 | | lysozyme (1sec) and UV lasr excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CIK
 
 | | Insights Into Crossreactivity in Human Allorecognition: The Structure of HLA-B35011 Presenting an Epitope derived from Cytochrome P450. | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-35 ALPHA CHAIN, ... | | Authors: | Hourigan, C.S, Harkiolaki, M, Peterson, N.A, Bell, J.I, Jones, E.Y, O'Callaghan, C.A. | | Deposit date: | 2006-03-22 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Eur.J.Immunol., 36, 2006
|
|
3ZVB
 
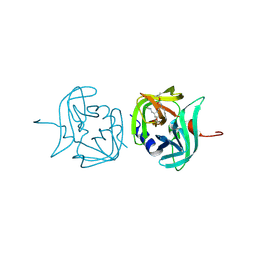 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 81 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVE
 
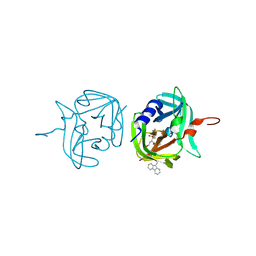 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | Descriptor: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
1OEG
 
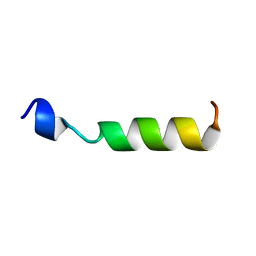 | | PEPTIDE OF HUMAN APOE RESIDUES 267-289, NMR, 5 STRUCTURES AT PH 6.0, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
2CKJ
 
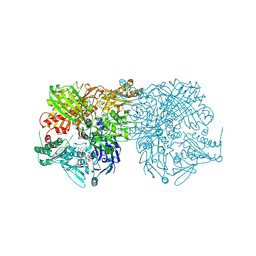 | | Human milk xanthine oxidoreductase | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pearson, A.R, Godber, B.L.J, Eisenthal, R, Taylor, G.L, Harrison, R. | | Deposit date: | 2006-04-19 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Human Milk Xanthine Dehydrogenase is Incompletely Converted to the Oxidase Form in the Absence of Proteolysis. A Structural Explanation.
To be Published
|
|
2CNH
 
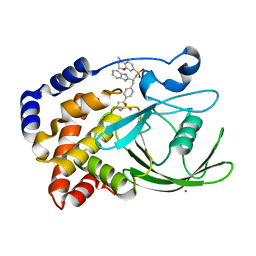 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | CALCIUM ION, N-[(1S)-1-(1H-BENZIMIDAZOL-2-YL)-2-{4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]PHENYL}ETHYL]-4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-SULFONAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
1O5C
 
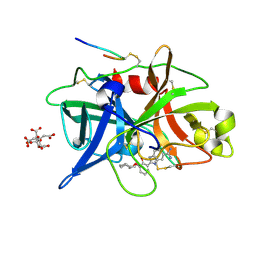 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
2CMC
 
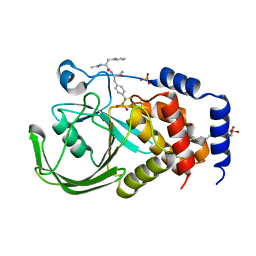 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-ACETYL-L-PHENYLALANYL-4-[DIFLUORO(PHOSPHONO)METHYL]-L-PHENYLALANINAMIDE, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, ... | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2CNF
 
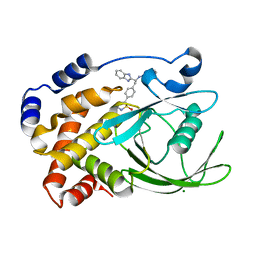 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | (5S)-5-{4-[(2S)-2-(1H-BENZIMIDAZOL-2-YL)-2-(1,3-BENZOTHIAZOL-2-YLAMINO)ETHYL]PHENYL}ISOTHIAZOLIDIN-3-ONE 1,1-DIOXIDE, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
4B6W
 
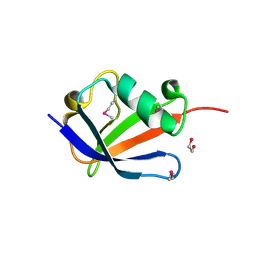 | | Architecture of Trypanosoma brucei Tubulin-Binding cofactor B | | Descriptor: | 1,2-ETHANEDIOL, TUBULIN-SPECIFIC CHAPERONE | | Authors: | Fleming, J.R, Morgan, R.E, Fyfe, P.K, Kelly, S.M, Hunter, W.N. | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-22 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Architecture of Trypanosoma Brucei Tubulin-Binding Cofactor B and Implications for Function.
FEBS J., 280, 2013
|
|
1RRE
 
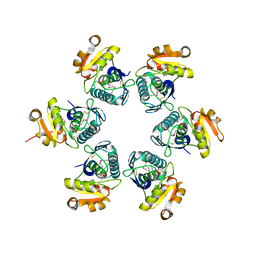 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1R4D
 
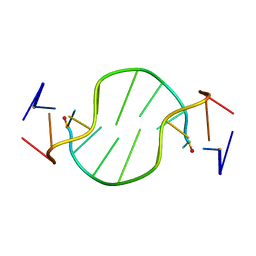 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
4S0O
 
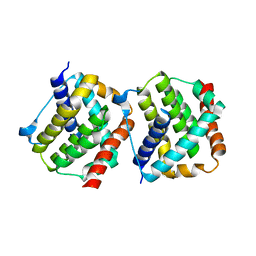 | |
1R88
 
 | | The crystal structure of Mycobacterium tuberculosis MPT51 (FbpC1) | | Descriptor: | MPT51/MPB51 antigen | | Authors: | Wilson, R.A, Maughan, W.N, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic alpha/beta hydrolases.
J.Mol.Biol., 335, 2004
|
|
