7SYS
 
 | | Structure of the delta dII IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(delta dII). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7U3E
 
 | | GID4 in complex with compound 1 | | Descriptor: | Glucose-induced degradation protein 4 homolog, tert-butyl (1S,4S)-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
5W0T
 
 | |
9FLR
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU1963 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-4-yl)-3-pyridin-4-yl-1~{H}-pyrazolo[1,5-a]pyrimidine, NICKEL (II) ION, ... | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8SSU
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
6UBO
 
 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
6UBV
 
 | |
7SYR
 
 | | Structure of the wt IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(wt). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8YH5
 
 | | A3R-Gi complex bound to i6A | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hemagglutinin,Adenosine receptor A3,GFP chimera, ... | | Authors: | Oshima, H.S, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
4SGA
 
 | |
7YFJ
 
 | | Crystal structure of human WTAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pre-mRNA-splicing regulator WTAP | | Authors: | Yan, X.H, Guan, Z.Y, Tang, C, Yin, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AI-empowered integrative structural characterization of m 6 A methyltransferase complex.
Cell Res., 32, 2022
|
|
6GFG
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5HWJ
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase | | Descriptor: | FORMIC ACID, GLYCEROL, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
4TYO
 
 | | PPIase in complex with a non-phosphate small molecule inhibitor. | | Descriptor: | 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-D-alanine, GLYCEROL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5KL3
 
 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
8CCA
 
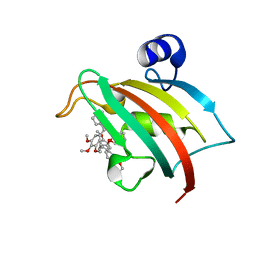 | | The Fk1 domain of FKBP51 in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
5KLX
 
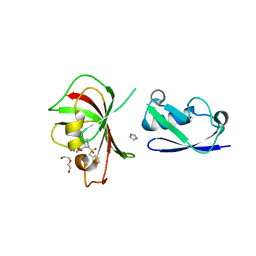 | |
5HKZ
 
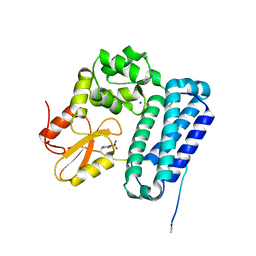 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
8CBH
 
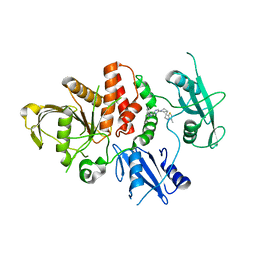 | | SHP2 in complex with a novel allosteric inhibitor | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [(1~{S},6~{R},7~{S})-3-[3-[2,3-bis(chloranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyrazin-6-yl]-7-(4-methyl-1,3-thiazol-2-yl)-3-azabicyclo[4.1.0]heptan-7-yl]methanamine | | Authors: | di Fabio, R, Petrocchi, A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
9GPN
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 11b/j | | Descriptor: | 3-cyclohexyl-1,19-dimethoxy-11,16-dioxa-19lambda4-polona-1,13-diaza-2lambda5-polonapentacyclo[16.4.0.04,9.013,19.020,25]docosane-2,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
4TVG
 
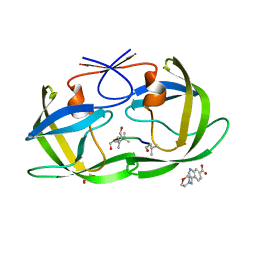 | | HIV Protease (PR) dimer in closed form with pepstatin in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, GLYCEROL, HIV Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
4TWI
 
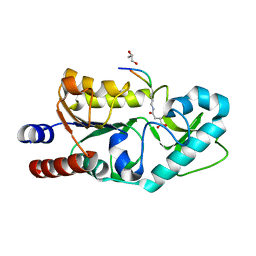 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
8C1N
 
 | |
7QHP
 
 | |
