8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | Descriptor: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | Authors: | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
9JKC
 
 | |
1NG6
 
 | | Structure of Cytosolic Protein of Unknown Function YqeY from Bacillus subtilis | | Descriptor: | Hypothetical protein yqeY | | Authors: | Zhang, R, Dementiva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4A crystal structure of hypothetical cytosolic protein
YQEY
To be Published
|
|
7AZO
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7MP6
 
 | | Neurofibromin homodimer | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9E0Q
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei D-hydrazino-Lysine analog at 2.3 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
8Z1R
 
 | | isocitrate lyase MoMcl1 | | Descriptor: | 1,2-ETHANEDIOL, Isocitrate lyase, MAGNESIUM ION | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | High-resolution crystal structure of the MoMcl1 protein
To Be Published
|
|
6O2A
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NDN and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
8FTJ
 
 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
8FLT
 
 | | Human PTH1R in complex with M-PTH(1-14) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
7B6T
 
 | | Sheep Polyomavirus VP1 in complex with 10 mM globo-N-tetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
6FH6
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-(2-azanylethyl)-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
7AVW
 
 | |
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8ZFK
 
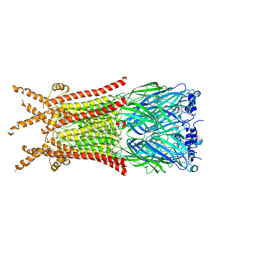 | | Caenorhabditis elegans ACR-23 in betaine and monepantel bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562, ... | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural insights into the molecular effects of the anthelmintics monepantel and betaine on the Caenorhabditis elegans acetylcholine receptor ACR-23.
Embo J., 43, 2024
|
|
6XBW
 
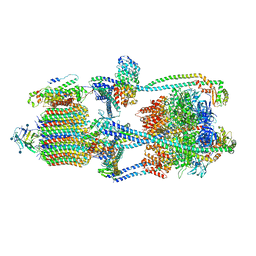 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
8CVS
 
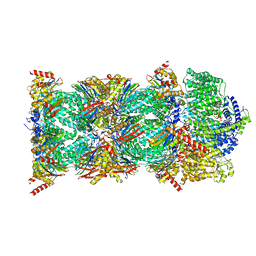 | | Human PA200-20S proteasome with MG-132 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome activator complex subunit 4, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
4ZTQ
 
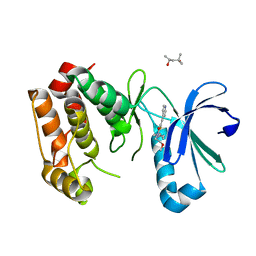 | | Human Aurora A catalytic domain bound to FK932 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-(2-methoxyethyl)-5-(pyridin-4-ylmethylidene)-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
7VHL
 
 | | Double deletion S-2P trimer(1 Up) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Cui, Z. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Double deletion S-2P trimer(1 Up)
To Be Published
|
|
8C00
 
 | |
3FAR
 
 | |
5LMR
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
4RX1
 
 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | Descriptor: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
5LP0
 
 | |
