8EID
 
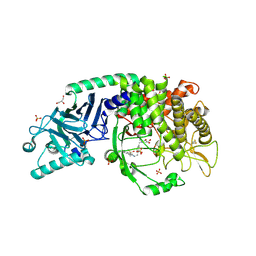 | | Co-crystal structure of Chaetomium glucosidase with compound 14 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({[(5P)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ELE
 
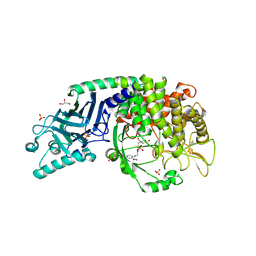 | | Co-crystal structure of Chaetomium glucosidase with compound 16 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-23 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4YAC
 
 | | Crystal structure of LigO in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
5HMH
 
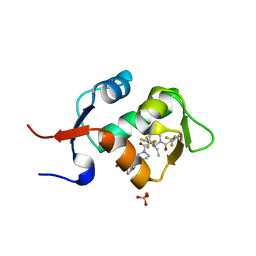 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | 4-[2-(4-{[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]carbonyl}piperazin-1-yl)phenoxy]butanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
6QRG
 
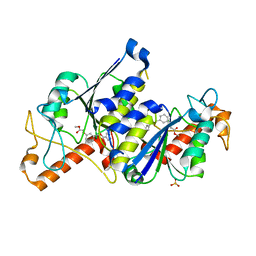 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | SULFATE ION, [3-(2-methoxy-2-oxidanylidene-ethoxy)phenyl]methyl 1~{H}-pyrazole-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
8UIY
 
 | | In situ human P-Z state 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wei, Z, Yong, X. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | In situ human P-Z state 80S ribosome
To Be Published
|
|
8PBN
 
 | | Mutant R1789Q of the dihydroorotase domain of human CAD protein bound to the inhibitor fluoorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | del Cano-Ochoa, F, Ramon-Maiques, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Beyond genetics: Deciphering the impact of missense variants in CAD deficiency.
J Inherit Metab Dis, 46, 2023
|
|
8BOU
 
 | | Crystal structure of Blautia producta GH94 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Blautia producta is a competent degrader among human gut Firmicutes for utilizing dietary beta mixed linkage glucan
To Be Published
|
|
8BK2
 
 | | X-ray structure of meningococcal factor H binding protein variant 2 in complex with a specific and bactericidal human monoclonal antibody 1B1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Veggi, D, Bottomley, J.M. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bactericidal human monoclonal antibody 1B1 shows specificity for meningococcal factor H binding protein variant 2 and displaces human factor H.
Faseb Bioadv, 6, 2024
|
|
8EUR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8F17
 
 | | Structure of the STUB1 TPR domain in complex with H204, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8PBK
 
 | | Mutant R1722W of the dihydroorotase domain of human CAD protein bound to the inhibitor fluoorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | del Cano-Ochoa, F, Ramon-Maiques, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Beyond genetics: Deciphering the impact of missense variants in CAD deficiency.
J Inherit Metab Dis, 46, 2023
|
|
6GNY
 
 | | Crystal structure of the MAJIN-TERB2 heterotetrameric complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Membrane-anchored junction protein, Telomere repeats-binding bouquet formation protein 2 | | Authors: | Dunce, J.M, Gurusaran, M, Sen, L.T, Davies, O.R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1.
Nat Commun, 9, 2018
|
|
4TKG
 
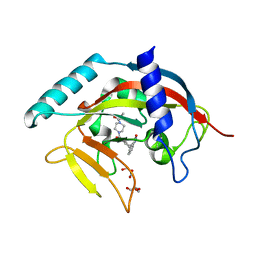 | | Crystal Structure of human Tankyrase 2 in complex with AZD2281. | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6Q7B
 
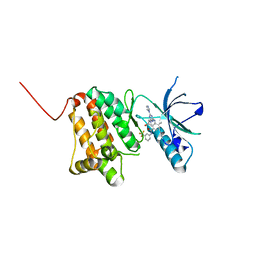 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL09 | | Descriptor: | 3-[[4-imidazol-1-yl-6-[(3~{S})-3-oxidanylpyrrolidin-1-yl]-1,3,5-triazin-2-yl]amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
9HMT
 
 | |
8GZ5
 
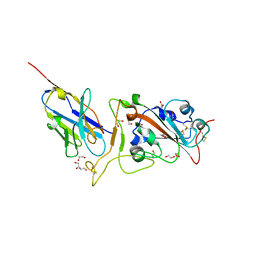 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
5MI6
 
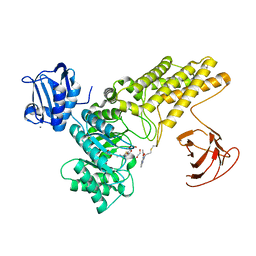 | | BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
6CPA
 
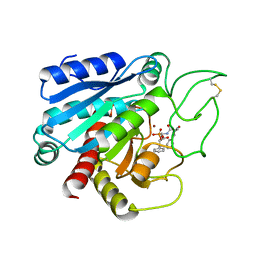 | |
8EQ7
 
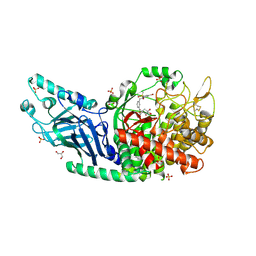 | | Co-crystal structure of Chaetomium glucosidase with compound 20 | | Descriptor: | (2R,3R,4R,5S)-1-[(3-{[3-bromo-5-(methanesulfonyl)anilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7TQ2
 
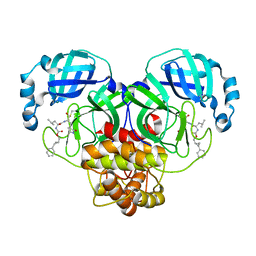 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8ER4
 
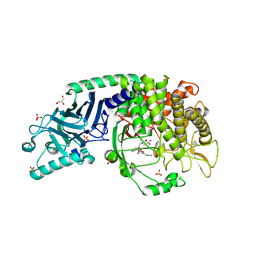 | | Co-crystal structure of Chaetomium glucosidase with compound 23 | | Descriptor: | (2R,3R,4R,5S)-1-(2-{6-[2-(4-azido-2-nitroanilino)ethyl]pyrazin-2-yl}ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
9H9O
 
 | |
7TQ3
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9MXW
 
 | |
