5WIY
 
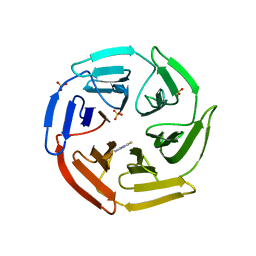 | | Kelch domain of human Keap1 bound to small molecule inhibitor fragment: 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5W97
 
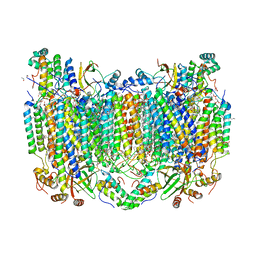 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Serial Femtosecond X-Ray Crystallography at Room Temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I, Zatsepin, N.A, Grant, T.D, Fromme, P, Fromme, R. | | Deposit date: | 2017-06-22 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3QTC
 
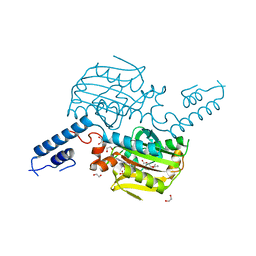 | | Crystal structure of the catalytic domain of MmOmeRS, an O-methyl tyrosyl-tRNA synthetase evolved from Methanosarcina mazei PylRS, complexed with O-methyl tyrosine and AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, O-methyl-L-tyrosine, ... | | Authors: | Dellas, N, Takimoto, J.K, Noel, J.P, Wang, L. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in Vivo Incorporation of Structurally Divergent Non-native Amino Acids.
Acs Chem.Biol., 6, 2011
|
|
3T4G
 
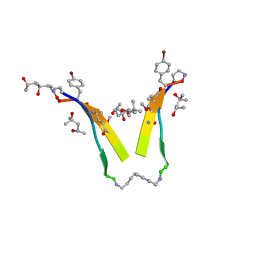 | | AIIGLMV segment from Alzheimer's Amyloid-Beta displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide (ORN)AIIGLMV(ORN)KF(HAO)(4BF)K | | Authors: | Zhao, M, Liu, C, Cheng, P.N, Eisenberg, D, Nowick, J.S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amyloid beta-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity.
Nat Chem, 4, 2012
|
|
2RM4
 
 | |
1SN6
 
 | |
9F1J
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[2-[2-[2-[2-[2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethanoylamino]ethoxy]ethoxy]ethoxy]ethyl]-4-[4-[2-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(2-morpholin-4-ylethyl)benzimidazol-2-yl]ethyl]phenoxy]butanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.13003039 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14
to be published
|
|
9F1N
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46 | | Descriptor: | (2S,4R)-1-[(2S)-2-[9-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]nonanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-4H-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.71000016 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46
to be published
|
|
9F1L
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[(3R)-2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethyl]-2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.30000627 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35
to be published
|
|
9F1M
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[7-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]heptanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.90001261 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45
to be published
|
|
9F1K
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]-N-(2-methoxyethyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30
to be published
|
|
7SPO
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with VNAR 3B4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2021-11-02 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography.
Nat Commun, 12, 2021
|
|
7SPP
 
 | |
1PG9
 
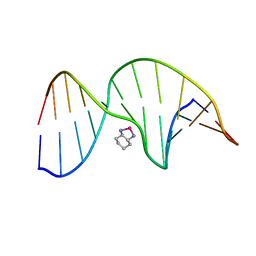 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
7T4E
 
 | |
7SXM
 
 | |
6X3Y
 
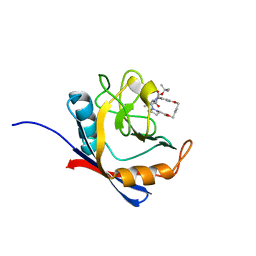 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4N
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 24) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
5BRW
 
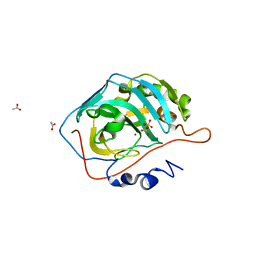 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BJZ
 
 | |
4V72
 
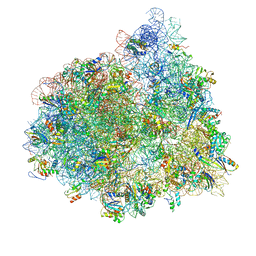 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6X3R
 
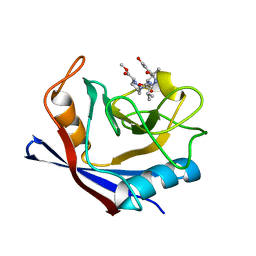 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, methyl (3~{S})-1-[(2~{S})-2-[[(2~{S})-2-acetamido-3-methyl-butanoyl]amino]-3-(3-hydroxyphenyl)propanoyl]-1,2-diazinane-3-carboxylate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4P
 
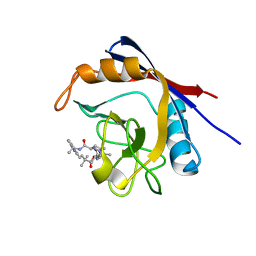 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 28) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
4V76
 
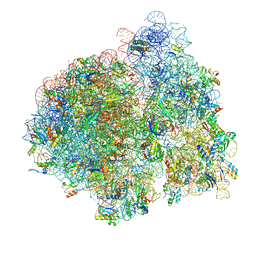 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post2a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
