4EVV
 
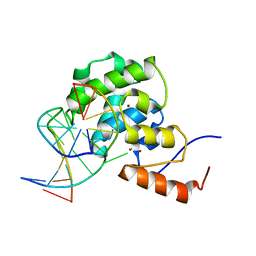 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4I6S
 
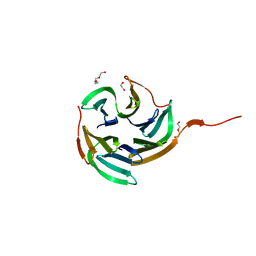 | | Structure of RSL mutant W76A in complex with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, Putative fucose-binding lectin protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Audfray, A, Arnaud, J, Varrot, A, Imberty, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Reduction of lectin valency drastically changes glycolipid dynamics in membranes but not surface avidity
Acs Chem.Biol., 8, 2013
|
|
2LZK
 
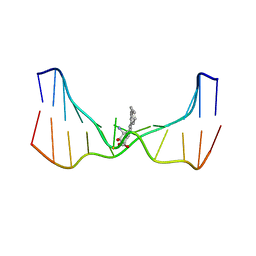 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
6PGX
 
 | |
6WNK
 
 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
5L8N
 
 | | crystal structure of human FABP6 protein with fragment 1 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 5,6-dimethyl-1~{H}-benzimidazol-2-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
6D6D
 
 | |
3HAX
 
 | |
5Z0D
 
 | |
6D6O
 
 | |
7A5X
 
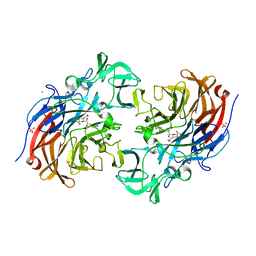 | | Two copies of the catalytic domain of NanA sialidase from Streptococcus pneumoniae juxtaposed in the P212121 space group, in complex with DANA derivatized with a PEG linker on the glycerol group. | | Descriptor: | (4~{S},5~{R},6~{R})-5-acetamido-6-[(1~{R},2~{S})-3-[[1-(2-ethoxyethyl)-1,2,3-triazol-4-yl]methylsulfanyl]-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bridot, C, Bouckaert, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polyvalent Transition-State Analogues of Sialyl Substrates Strongly Inhibit Bacterial Sialidases*.
Chemistry, 27, 2021
|
|
1G4Q
 
 | | RNA/DNA HYBRID DECAMER OF CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', ... | | Authors: | Han, G.W. | | Deposit date: | 2000-10-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct-methods determination of an RNA/DNA hybrid decamer at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
9F0R
 
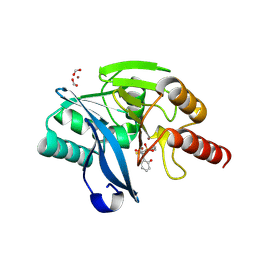 | | VIM-2 in complex with GKV65 (5g) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Bosman, R, Prester, A, Bartels, K, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
6MI2
 
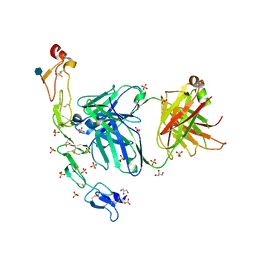 | | Structure of the human 4-1BB / Utomilumab Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
8IOT
 
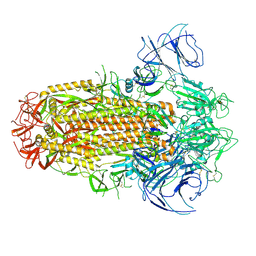 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
5LBX
 
 | | Structure of the T175V Etr1p mutant in the trigonal form P312 in complex with NADP and crotonyl-CoA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
6FPN
 
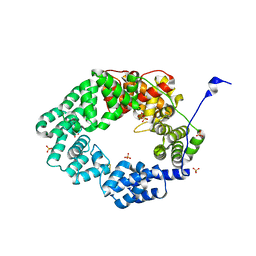 | | Lytic transglycosylase in action | | Descriptor: | Putative soluble lytic murein transglycosylase, SULFATE ION | | Authors: | Williams, A.H, Hoauz, A, Boneca, I.G. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A step-by-stepin crystalloguide to bond cleavage and 1,6-anhydro-sugar product synthesis by a peptidoglycan-degrading lytic transglycosylase.
J. Biol. Chem., 293, 2018
|
|
5F6Y
 
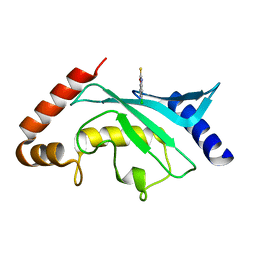 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6P0W
 
 | |
6DB8
 
 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | 2-[(Z)-(3-methyl-1,3-benzoxazol-2(3H)-ylidene)methyl]-3-(3-sulfopropyl)-1,3-benzothiazol-3-ium, Fab-Heavy chain, Fab-Light chain, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86541343 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
8W71
 
 | | Structural basis of chorismate isomerization by Arabidopsis isochorismate synthase ICS1 | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, FORMIC ACID, Isochorismate synthase 1, ... | | Authors: | Su, Z.H, Ming, Z.H. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural basis of chorismate isomerization by Arabidopsis ISOCHORISMATE SYNTHASE1.
Plant Physiol., 196, 2024
|
|
5ZH3
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-6 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, LYSINE, Lysine-tRNA ligase | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
8A8D
 
 | |
6D6B
 
 | |
8A7J
 
 | | PcIDS1 in complex with Mn2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
