9F22
 
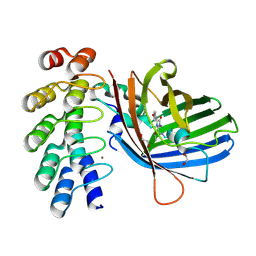 | | DARPin eGFP complex DP1 (3G190.24) | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DARPin DP1, ... | | Authors: | Mittl, P.R, Winkelvoss, D. | | Deposit date: | 2024-04-22 | | Release date: | 2024-10-23 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular features defining the efficiency of bioPROTACs.
Commun Biol, 8, 2025
|
|
6IKX
 
 | |
9FMZ
 
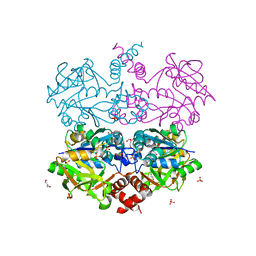
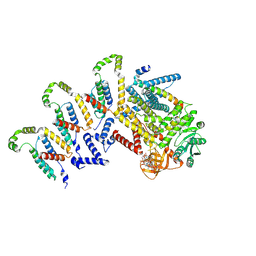 | | Cryo-EM structure of the c-di-GMP-bound synthase:pEtN transferase complex (BcsA-Bct-G3) from the E. coli cellulose secretion macrocomplex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cellulose biosynthesis protein BcsG, Cellulose synthase catalytic subunit [UDP-forming], ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
6Z1C
 
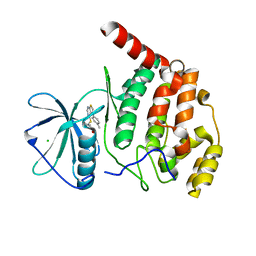 | | Crystal structure of Arabidopsis thaliana CK2-alpha-1 in complex with TTP-22 | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha-1 | | Authors: | Demulder, M, Loris, R, De Veylder, L. | | Deposit date: | 2020-05-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway.
Plant Cell, 33, 2021
|
|
9BP8
 
 | | Crystal structure of BRAF kinase domain with PF-07799933 | | Descriptor: | DIMETHYL SULFOXIDE, N-{(1R,2P)-6-chloro-5-[(5-chloro-3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-fluorocyclohexa-2,3,5-trien-1-yl}-3-fluoroazetidine-1-sulfonamide, SULFATE ION, ... | | Authors: | Mou, T.-C. | | Deposit date: | 2024-05-07 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of BRAF kinase domain with PF-07799933
To Be Published
|
|
9J19
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 3C-like proteinase nsp5 | | Authors: | Singh, A, Jangid, K, Dhaka, P, Tomar, S, Kumar, P. | | Deposit date: | 2024-08-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into the Main Protease (Mpro) Dimer Interface Destabilization Inhibitor: Unveiling New Therapeutic Avenues against SARS-CoV-2.
Biochemistry, 64, 2025
|
|
8EY0
 
 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
5FFV
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with H3K14ac histone peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3, Peregrin | | Authors: | Tallant, C, Nunez-Alonso, G, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2015-12-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with H3K14ac histone peptide
To Be Published
|
|
5KL7
 
 | | Wilms Tumor Protein (WT1) ZnF2-4Q369R in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
6QF2
 
 | | X-Ray structure of Thermolysin crystallized on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6INE
 
 | | Crystal Structure of human ASH1L-MRG15 complex | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase ASH1L, Mortality factor 4-like protein 1, ... | | Authors: | Hou, P, Huang, C, Liu, C.P, Yu, T, Yin, Y, Zhu, B, Xu, R.M. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Stimulation of Ash1L's H3K36 Methyltransferase Activity through Mrg15 Binding.
Structure, 27, 2019
|
|
9H3J
 
 | |
9HLJ
 
 | | Crystal structure of GV37-TCR in complex with HLA-C*12:02 with KAYNVTQAF (KF9), a 9-mer epitope from SARS-CoV-2 Nucleocapsid (N266-274) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GV37-TCR alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Chatzileontiadou, D.S.M, Gras, S. | | Deposit date: | 2024-12-05 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis of potent antiviral HLA-C-restricted CD8 + T cell response to an immunodominant SARS-CoV-2 nucleocapsid epitope.
Nat Commun, 16, 2025
|
|
5LRE
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
4TPT
 
 | | Crystal Structure of the Human LIMK2 Kinase Domain In Complex With a Non-ATP Competitive Inhibitor | | Descriptor: | LIM domain kinase 2, N-{4-[(1S)-1,2-dihydroxyethyl]benzyl}-N-methyl-4-(phenylsulfamoyl)benzamide | | Authors: | Goodwin, N.C, Cianchetta, G, Hamman, B.L, Burgoon, H.A, Healy, J, Mabon, S, Strobel, E.D, Wang, S, Rawlins, D.B. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Type III Inhibitor of LIM Kinase 2 That Binds in a DFG-Out Conformation.
Acs Med.Chem.Lett., 6, 2015
|
|
6QDA
 
 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000811 | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
8F9X
 
 | | Cyclase-Phosphotriesterase from Ruegeria pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Frkic, R.L, Ji, D, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-06 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Thermostable Bacterial Metallohydrolase that Degrades Organophosphate Plasticizers.
Chembiochem, 26, 2025
|
|
8CA7
 
 | | Omadacycline and spectinomycin bound to the 30S ribosomal subunit head | | Descriptor: | 16S rRNA, 30S ribosomal protein S7, MAGNESIUM ION, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7W10
 
 | | UGT74AN2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Long, F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
6QFG
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK144 | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5IAZ
 
 | |
5UXV
 
 | |
6I2S
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD (R802A) in complex with GarA, following 2-oxoglutarate soak | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, CALCIUM ION, Glycogen accumulation regulator GarA, ... | | Authors: | Wagner, T, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling.
Sci.Signal., 12, 2019
|
|
6I76
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-3 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azido-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
