8UW2
 
 | | Structure of AKT1(E17K) with compound 3 (zinc-free) | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
9GPX
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/l | | Descriptor: | 3-cyclohexyl-17-hydroxy-16,17-dimethoxy-12-methyl-15,18-dioxa-5-aza-1lambda5,17lambda5-diphosphapentacyclo[17.3.1.01,10.05,10.011,19]tricosa-10,19(23)-diene-4,11-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5KDV
 
 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, SULFATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6NW3
 
 | | BACE1 in complex with a macrocyclic inhibitor | | Descriptor: | Beta-secretase 1, N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, SULFATE ION | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-02-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
9GPL
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 11c/i | | Descriptor: | 2-cyclohexyl-1,19-dimethoxy-11,18-dioxa-1lambda5,19lambda5-diphospha-2-stannatetracyclo[15.3.0.01,13.04,9]icosane-3,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPW
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/j | | Descriptor: | 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7Z0J
 
 | | human PEX13 SH3 domain in complex with internal FxxxF motif | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13 | | Authors: | Gaussmann, S, Zak, K, Kreisz, N, Sattler, M. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of peroxisomal import by the PEX13 SH3 domain and a proximal FxxxF binding motif.
Nat Commun, 15, 2024
|
|
6FFZ
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H, Y54L | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
8UW7
 
 | | Structure of AKT1(WT) with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Mutant-selective AKT inhibition through lysine targeting and neo-zinc chelation.
Nature, 637, 2025
|
|
8WWS
 
 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
9GQ5
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,2)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-20,23-dimethoxy-11,18-dioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(22),14,19(23),20-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
4RE4
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
8GG5
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #11 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG6
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #12 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7Z6J
 
 | |
9GQ8
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,2)-H2 | | Descriptor: | (2~{S},9~{S})-2-cyclohexyl-20,23-dimethoxy-11,18-dioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(21),19,22-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8GG7
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #13 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8BS9
 
 | | Structure of USP36 in complex with Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 36, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
9GQ9
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,2)-(Z) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-20,23-dimethoxy-11,18-dioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(22),14,19(23),20-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQA
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,2)-Diol-IV | | Descriptor: | (2~{S},9~{S},14~{S},15~{R})-2-cyclohexyl-20,23-dimethoxy-14,15-bis(oxidanyl)-11,18-dioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(21),19,22-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7YSI
 
 | | Crystal structure of thioredoxin 2 | | Descriptor: | Thiol disulfide reductase thioredoxin, ZINC ION | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Comparison of the structure and activity of thioredoxin 2 and thioredoxin 1 from Acinetobacter baumannii.
Iucrj, 10, 2023
|
|
8GG8
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #14 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG9
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #15 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
4FQH
 
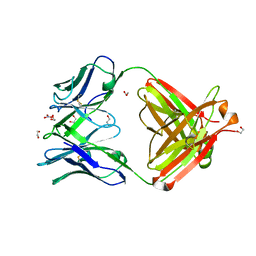 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
8GGB
 
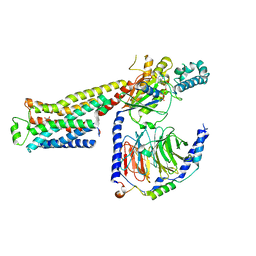 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #17 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
