7ZI8
 
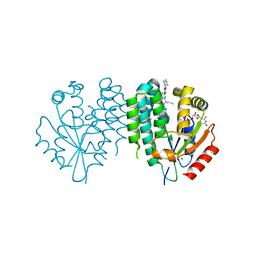 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0602 inhibitor | | Descriptor: | 2-[2-[[2-methyl-5-[6-[2-(4-methylpiperazin-1-yl)ethyl]pyridin-3-yl]phenyl]-propyl-amino]-1,3-thiazol-4-yl]pyrimidine-4,6-diamine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
7PE8
 
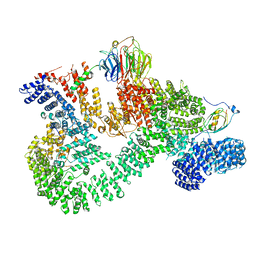 | |
6MR8
 
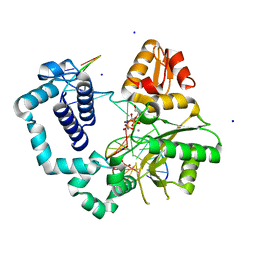 | | D276G DNA polymerase beta substrate complex with templating adenine and incoming Fapy-dGTP analog | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
6ZXM
 
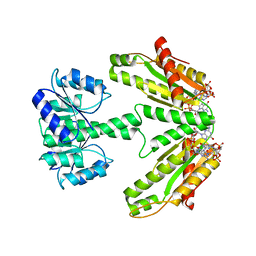 | | Diguanylate cyclase DgcR in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Putative GGDEF/response regulator receiver domain protein | | Authors: | Teixeira, R.D, Schirmer, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Activation mechanism of a small prototypic Rec-GGDEF diguanylate cyclase.
Nat Commun, 12, 2021
|
|
8TXN
 
 | |
6ZMD
 
 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
8RHS
 
 | |
4Q19
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 5 {5-(4-{[(4,6-DIAMINOPYRIMIDIN-2-YL)SULFANYL]METHYL}-5-PROPYL-1,3-THIAZOL-2-YL)-2-METHOXYPHENOL} | | Descriptor: | 5-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
7PE7
 
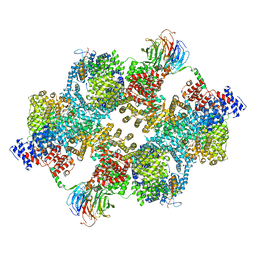 | |
7A4D
 
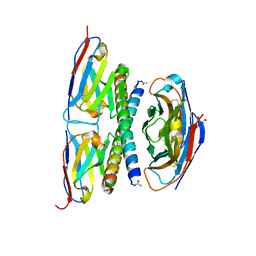 | |
8HY6
 
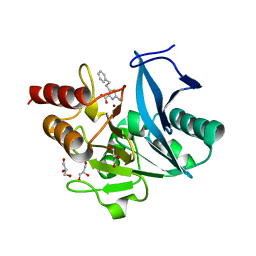 | | Crystal structure of B1 NDM-1 MBL in complex with 2-amino-5-phenethylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-phenylethyl)-1,3-thiazole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
9CF7
 
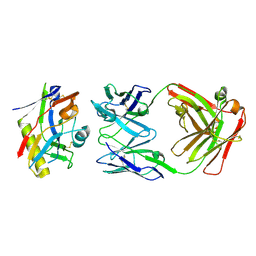 | |
8TS5
 
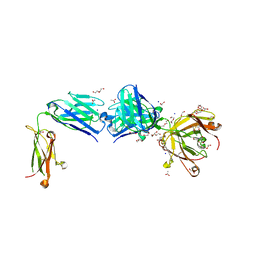 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
6XYR
 
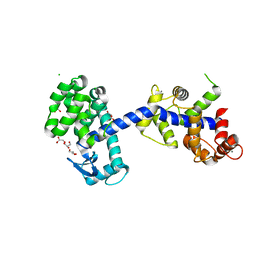 | | Structure of the T4Lnano fusion protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
6MRG
 
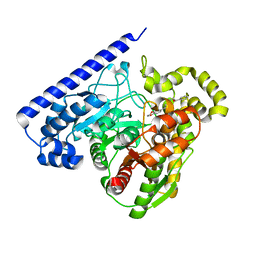 | | FAAH bound to non covalent inhibitor | | Descriptor: | (1R)-2-{[6-(2,3-dihydro-1,4-benzodioxin-6-yl)pyrimidin-4-yl]amino}-1-phenylethan-1-ol, Fatty-acid amide hydrolase 1 | | Authors: | Saha, A, Shih, A, Mirzadegan, T, Seierstad, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Predicting the Binding of Fatty Acid Amide Hydrolase Inhibitors by Free Energy Perturbation.
J Chem Theory Comput, 14, 2018
|
|
7X1W
 
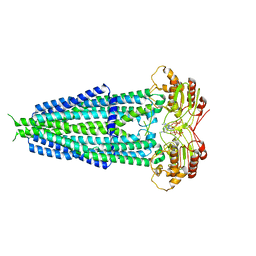 | |
6XZR
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 1 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6S11
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 6-pyridin-4-yl-3-[3-(trifluoromethyloxy)phenyl]imidazo[1,2-b]pyridazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
8A51
 
 | | Crystal structure of HSF2BP-BRME1 complex | | Descriptor: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8P0F
 
 | | Crystal structure of the VCB complex with compound 1. | | Descriptor: | (3~{R},5~{R})-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-5-oxidanyl-2-oxidanylidene-1-pyridin-2-yl-piperidine-3-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Drugit: crowd-sourcing molecular design of non-peptidic VHL binders.
Nat Commun, 16, 2025
|
|
6GBS
 
 | | Crystal Structure of the C. themophilum Scavenger Decapping Enzyme DcpS apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putative mRNA decapping protein | | Authors: | Fuchs, A.-L, Neu, A, Sprangers, R. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Molecular basis of the selective processing of short mRNA substrates by the DcpS mRNA decapping enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8QY2
 
 | | Xylanase from Bacillus circulans mutant E78Q/F125A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX.
Febs J., 291, 2024
|
|
6PF2
 
 | | Crystal Structure of Amino Acids 1220-1276 of Human Beta Cardiac Myosin Fused to Gp7 and Eb1 | | Descriptor: | 1,2-ETHANEDIOL, Myosin, heavy polypeptide 7, ... | | Authors: | Andreas, M.P, Korkmaz, E.N, Kirsch, C.J, Hargreaves, M, Kieffer, D.J, Ajay, G, Cui, Q, Rayment, I. | | Deposit date: | 2019-06-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Complete Model of the Cardiac Myosin Rod
To Be Published
|
|
8HWT
 
 | | SARS-CoV-2 Omicron BA.2 RBD complexed with BD-604 and S304 Fab | | Descriptor: | BD-604 heavy chain, BD-604 light chain, S304 heavy chain, ... | | Authors: | He, Q.W, Xie, Y. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
7ZL8
 
 | | NME1 in complex with succinyl-CoA | | Descriptor: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
