2W6X
 
 | | Crystal structure of Sperm Whale Myoglobin mutant YQRF in complex with Xenon | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | When the Same Fold Does not Mean the Same Function: The Case of Xenon Cavities in Hemoglobin and Myoglobin
To be Published
|
|
2W6Y
 
 | | Crystal structure of Sperm Whale Myoglobin mutant YQR in complex with Xenon | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | When the Same Fold Does not Mean the Same Function: The Case of Xenon Cavities in Hemoglobin and Myoglobin
To be Published
|
|
3GB4
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Cobalt and Dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, COBALT (II) ION, DdmC, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-02-18 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
2PKQ
 
 | | Crystal structure of the photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, Glyceraldehyde-3-phosphate dehydrogenase B, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PKR
 
 | | Crystal structure of (A+CTE)4 chimeric form of photosyntetic glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase Aor, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3GTS
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Iron and Dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, DdmC, FE (III) ION, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-28 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
2F36
 
 | |
2F34
 
 | |
2F35
 
 | |
3GTE
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Iron | | Descriptor: | ACETATE ION, DdmC, FE (III) ION, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
1PXV
 
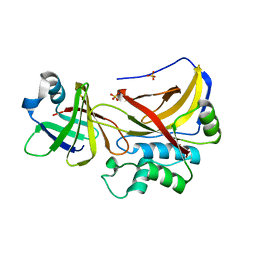 | | The staphostatin-staphopain complex: a forward binding inhibitor in complex with its target cysteine protease | | Descriptor: | GUANIDINE, SULFATE ION, cysteine protease, ... | | Authors: | Filipek, R, Rzychon, M, Oleksy, A, Gruca, M, Dubin, A, Potempa, J, Bochtler, M. | | Deposit date: | 2003-07-07 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Staphostatin-Staphopain Complex: A FORWARD BINDING INHIBITOR IN COMPLEX WITH ITS TARGET CYSTEINE PROTEASE.
J.Biol.Chem., 278, 2003
|
|
2FKP
 
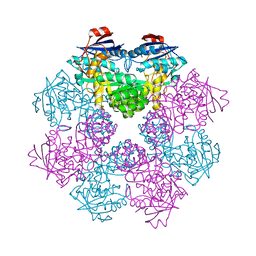 | |
3VEC
 
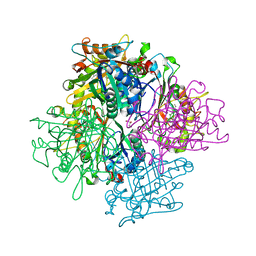 | | Rhodococcus jostii RHA1 DypB D153A variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, GLYCEROL, ... | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3VEF
 
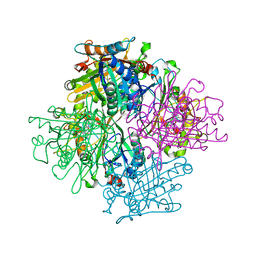 | | Rhodococcus jostii RHA1 DypB N246H variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3WOW
 
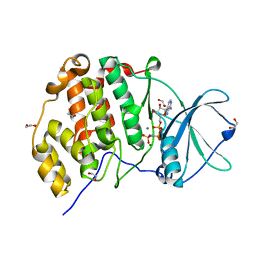 | | Crystal structure of human CK2a with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Sogabe, Y, Sakurai, A, Nakamura, S, Nakanishi, I, Shimada, K, Tanaka, M. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic residue divergence of CK2a contribute to a species-dependent variation for apigenin binding mode but not for an ATP analogue
To be Published
|
|
2IQ6
 
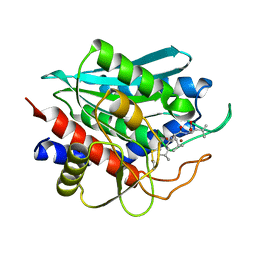 | | Crystal Structure of the Aminopeptidase from Vibrio proteolyticus in Complexation with Leucyl-leucyl-leucine. | | Descriptor: | Bacterial leucyl aminopeptidase, Peptide, (Leucyl-leucyl-leucine), ... | | Authors: | Kumar, A, Narayanan, B, Kim, J.-J.P, Bennett, B. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental evidence for a metallohydrolase mechanism in which the nucleophile is not delivered by a metal ion: EPR spectrokinetic and structural studies of aminopeptidase from Vibrio proteolyticus
Biochem.J., 403, 2007
|
|
4B7W
 
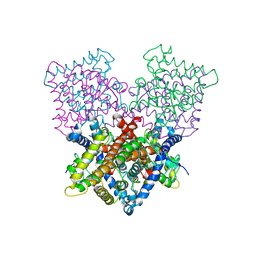 | | Ligand binding domain human hepatocyte nuclear factor 4alpha: Apo form | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 4-ALPHA | | Authors: | Dudasova, Z, Okvist, M, Kretova, M, Ondrovicova, G, Skrabana, R, LeGuevel, R, Salbert, G, Leonard, G, McSweeney, S, Barath, P. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Fatty Acids are not Essential Structural Components of Hepatocyte Nuclear Factor 4Alpha
To be Published
|
|
3EB8
 
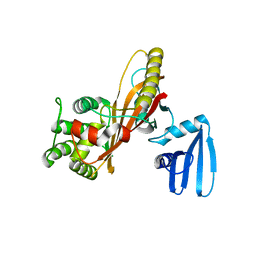 | | VirA | | Descriptor: | Cysteine protease-like virA | | Authors: | Germane, K.L, Spiller, B.W. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules.
Biochemistry, 47, 2008
|
|
2KCN
 
 | | Solution structure of the antifungal protein PAF from Penicillium chrysogenum | | Descriptor: | Antifungal protein | | Authors: | Batta, G, Barna, T, Gaspari, Z, Sandor, S, Kover, K.E, Binder, U, Sarg, B, Kaiserer, L, Chhillar, A.K, Eigentler, A, Leiter, E, Hegedus, N, Pocsi, I, Lindner, H, Marx, F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional aspects of the solution structure and dynamics of PAF--a highly-stable antifungal protein from Penicillium chrysogenum
Febs J., 276, 2009
|
|
2I0B
 
 | |
2I0C
 
 | |
2Q74
 
 | | Mycobacterium tuberculosis SuhB | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Brown, A.K, Meng, G, Ghadbane, H, Besra, G.S, Futterer, K. | | Deposit date: | 2007-06-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+
Bmc Struct.Biol., 7, 2007
|
|
3E76
 
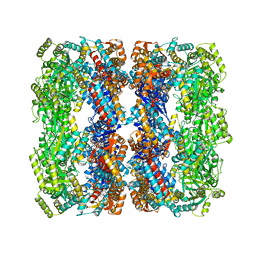 | | Crystal structure of Wild-type GroEL with bound Thallium ions | | Descriptor: | 60 kDa chaperonin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kiser, P.D, Lorimer, G.H, Palczewski, K. | | Deposit date: | 2008-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Use of thallium to identify monovalent cation binding sites in GroEL.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2QDX
 
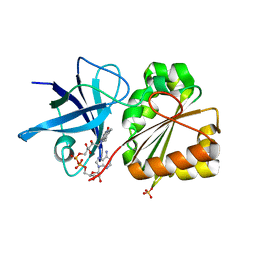 | | P.Aeruginosa Fpr with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase, SULFATE ION | | Authors: | Han, H, Schonbrunn, E. | | Deposit date: | 2007-06-21 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP(+) Reductase and Not Ferredoxin Is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions
Biochemistry, 46, 2007
|
|
1CQZ
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
