6YNM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
8PUN
 
 | | Old Yellow Enzyme from the thermophilic Ferrovum sp. JA12 | | Descriptor: | 1,2-ETHANEDIOL, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, CHLORIDE ION, ... | | Authors: | Polidori, N, Gruber, K. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Oligomerization, and Thermal Stability of a Recently Discovered Old Yellow Enzyme.
Proteins, 93, 2025
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
8GJL
 
 | | multi-drug efflux pump RE-CmeB bound with Ciprofloxacin | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-05-31 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
5GTT
 
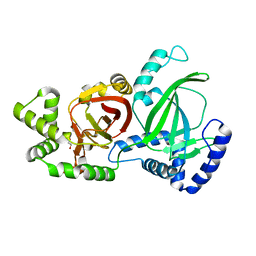 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | Descriptor: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5H4D
 
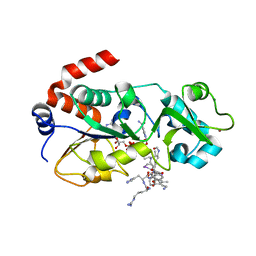 | | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 7-AMINO-4-METHYL-CHROMEN-2-ONE, ARG-HIS-LYS, ... | | Authors: | Zhang, S, Fu, L, Liu, J, Liu, B. | | Deposit date: | 2016-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride
To Be Published
|
|
6Q4V
 
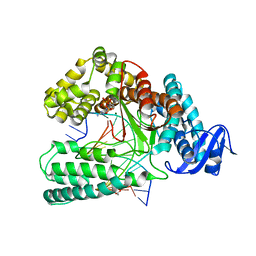 | | KlenTaq DNA polymerase in complex with dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6H57
 
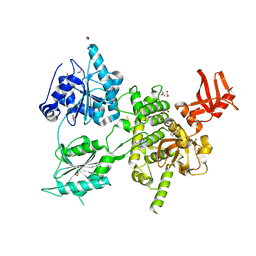 | | Crystal structure of S. cerevisiae DEAH-box RNA helicase Dhr1, essential for small ribosomal subunit biogenesis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Roychowdhury, A, Graille, M. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis.
Nucleic Acids Res., 47, 2019
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6Y73
 
 | | The crystal structure of human MACROD2 in space group P43 | | Descriptor: | ADP-ribose glycohydrolase MACROD2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8S0O
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3-[4-chloranyl-2-(1H-pyrazol-4-ylmethyl)indazol-5-yl]-5-methyl-6-(piperazin-1-ylmethyl)-1H-pyrrolo[3,2-b]pyridine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
6N5M
 
 | |
7B1R
 
 | |
6GR9
 
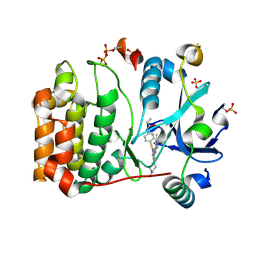 | | Human AURKC INCENP complex bound to VX-680 | | Descriptor: | Aurora kinase C, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Inner centromere protein, ... | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AURKC INCENP complex bound to BRD-7880
To Be Published
|
|
8AGM
 
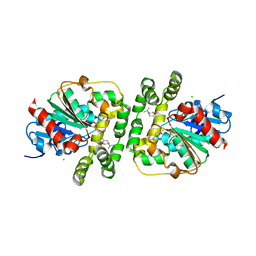 | | Limonene epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, CHLORIDE ION, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8TTR
 
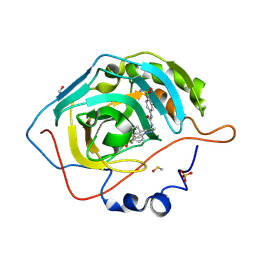 | | CA9 mimic bound to SH7 | | Descriptor: | 4-{5-phenyl-4-[(4-sulfamoylphenyl)carbamamido]-1H-pyrazol-1-yl}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2023-08-14 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 4-(Pyrazolyl)benzenesulfonamide Ureas as Carbonic Anhydrases Inhibitors and Hypoxia-Mediated Chemo-Sensitizing Agents in Colorectal Cancer Cells.
J.Med.Chem., 67, 2024
|
|
8JHO
 
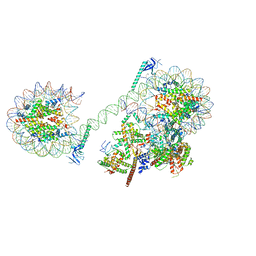 | |
8GRN
 
 | | AtOSCA1.1 extended state | | Descriptor: | Protein OSCA1, [1-MYRISTOYL-GLYCEROL-3-YL]PHOSPHONYLCHOLINE | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
7Q4R
 
 | | Crystal structure of human HSP72-NBD in complex with fragment 1 | | Descriptor: | 4-(4-phenyl-1,3-thiazol-2-yl)piperazine-1-carboxamide, Heat shock 70 kDa protein 1A, MAGNESIUM ION, ... | | Authors: | Le Bihan, Y.V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Characterization of a Cryptic Secondary Binding Site in the Molecular Chaperone HSP70.
Molecules, 27, 2022
|
|
6H5J
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 4 | | Descriptor: | (3~{R})-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Otero, J.M, Maneiro, M, Lence, E, Sanz-Gaitero, M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
9KKP
 
 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
8WG6
 
 | |
8JJM
 
 | |
