6T98
 
 | | Trypanothione Reductase from Leishmania infantum in complex with 9a | | Descriptor: | 4-[3-methyl-1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-3-ium-4-yl]butan-1-amine, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMD
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6HNB
 
 | | Crystal structure of aminotransferase Aro8 from Candida albicans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, CHLORIDE ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
8WOU
 
 | |
6Q5Z
 
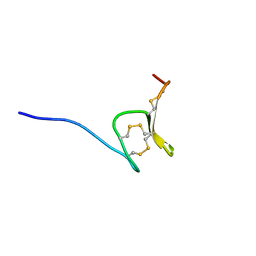 | | H-Vc7.2, H-superfamily conotoxin | | Descriptor: | Conotoxin Vc7.2 | | Authors: | Nielsen, L.D, Foged, M.M, Teilum, K, Ellgaard, L. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework.
J.Biol.Chem., 294, 2019
|
|
6I7N
 
 | |
8WKJ
 
 | |
6HNU
 
 | | Crystal structure of the aminotransferase Aro8 from C. Albicans with ligands | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
4GRX
 
 | | Structure of an omega-aminotransferase from Paracoccus denitrificans | | Descriptor: | Aminotransferase, DELTA-AMINO VALERIC ACID, SODIUM ION | | Authors: | Rausch, C, Lerchner, A, Schiefner, A, Skerra, A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the omega-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential.
Proteins, 81, 2013
|
|
6T95
 
 | | Trypanothione Reductase from Leismania infantum in complex with 4a | | Descriptor: | 1-[2-[5-[4-(4-azanylbutyl)-3-methyl-1,2,3-triazol-3-ium-1-yl]-2-[4-(2-phenylethyl)-1,3-thiazol-2-yl]phenoxy]ethyl]imidazolidin-2-one, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
6PPC
 
 | | Solution structure of conotoxin MiXXVIIA | | Descriptor: | Conopeptide phi-MiXXVIIA | | Authors: | Daly, N.L, Dekan, Z, Jin, A.H, Alewood, P.F. | | Deposit date: | 2019-07-06 | | Release date: | 2019-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conotoxin phi-MiXXVIIA from the Superfamily G2 Employs a Novel Cysteine Framework that Mimics Granulin and Displays Anti-Apoptotic Activity.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6JIF
 
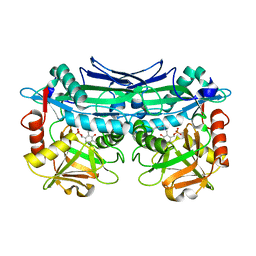 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
5GR0
 
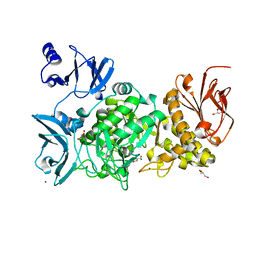 | |
5GR6
 
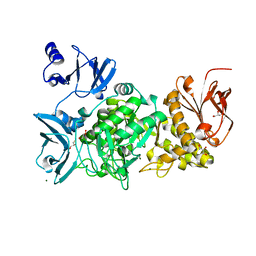 | |
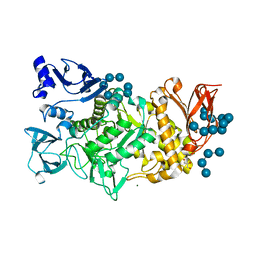
5GR4
 
 | |
5GQY
 
 | |
5GR1
 
 | |
5GR2
 
 | |
5GR5
 
 | |
5GQZ
 
 | |
7S5E
 
 | |
5EBK
 
 | |
5F8V
 
 | |
5GR3
 
 | |
