7D7X
 
 | |
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6XG1
 
 | | Class C beta-lactamase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
5JFR
 
 | | Potent, Reversible MetAP2 Inhibitors via Fragment Based Drug Discovery | | Descriptor: | 1,2-ETHANEDIOL, 7-fluoro-4-(5-methyl-3H-imidazo[4,5-b]pyridin-6-yl)-2,4-dihydropyrazolo[4,3-b]indole, DIMETHYL SULFOXIDE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 2.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7BHT
 
 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7STN
 
 | | Chitin Synthase 2 from Candida albicans bound to Nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), 1,2-Distearoyl-sn-glycerophosphoethanolamine, Chitin synthase | | Authors: | Ren, Z, Chhetri, A, Lee, S, Yokoyama, K. | | Deposit date: | 2021-11-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for inhibition and regulation of a chitin synthase from Candida albicans.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7D81
 
 | |
6XEB
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND E) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7D7W
 
 | |
8VOV
 
 | | Structure of VCP in complex with an ATPase activator and ADP (D2 domains only, hexameric form) | | Descriptor: | (3R)-N-[2-(ethylsulfanyl)phenyl]-3-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)butanamide, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Jones, N.H, Urnivicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric activation of VCP, an AAA unfoldase, by small molecule mimicry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
8PFO
 
 | | Crystal structure of WRN helicase domain in complex with HRO761 | | Descriptor: | Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION, ~{N}-[2-chloranyl-4-(trifluoromethyl)phenyl]-2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-6-[4-(6-methyl-5-oxidanyl-pyrimidin-4-yl)carbonylpiperazin-1-yl]-7-oxidanylidene-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]ethanamide | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
7D7Y
 
 | |
8ZSB
 
 | |
9AYM
 
 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9B0I
 
 | |
9B0E
 
 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-amide-CoA bisubstrate probe and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA cytidine acetyltransferase, [[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[[1-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-2-oxidanylidene-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
7QPC
 
 | | Inward-facing NPA bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
6O5T
 
 | | Crystal Structure of VIM-2 with Compound 16 | | Descriptor: | ACETATE ION, Beta-lactamase class B VIM-2, ZINC ION, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
5MGK
 
 | |
7QLP
 
 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8PPS
 
 | | Dimeric RbdA EAL, in apo state | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, EAL domain-containing protein, ... | | Authors: | Cordery, C.R, Maly, M, Walsh, M.A, Tews, I. | | Deposit date: | 2023-07-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Control of phosphodiesterase activity in the regulator of biofilm dispersal RbdA from Pseudomonas aeruginosa.
Rsc Chem Biol, 5, 2024
|
|
6O95
 
 | | Structure of the IRAK4 kinase domain with compound 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-(hydroxymethyl)-2-methyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
7SYF
 
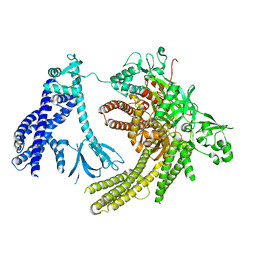 | | Reconstruction of full-length Prex-1 (PtdIns(3,4,5)P3-dependent Rac Exchanger 1) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein,Endolysin chimera | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-11-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8EN4
 
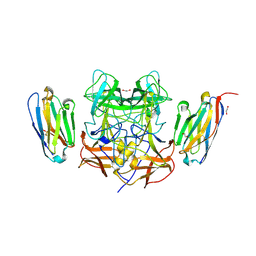 | | Structure of GII.4 norovirus in complex with Nanobody 53 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 53, VP1 | | Authors: | Kher, G, Sabin, C, Koromyslova, A, Pancera, M, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
