8JJY
 
 | | Crystal structure of QN-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8EDY
 
 | |
8JJW
 
 | | Crystal structure of QG-hNTAQ1 C28S | | Descriptor: | MAGNESIUM ION, Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJZ
 
 | | Crystal structure of QQ-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8G9Z
 
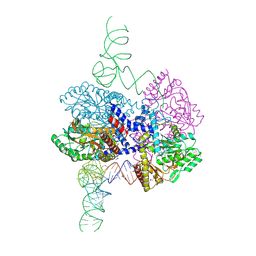 | | High-resolution crystal structure of the human selenomethionine-derived SepSecS-tRNASec complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Puppala, A, Simonovic, M, Castillo Suchkou, J. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 51, 2023
|
|
8JJX
 
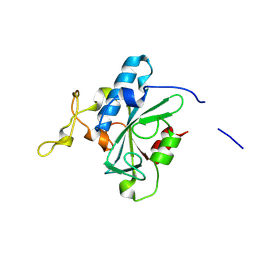 | | Crystal structure of QS-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
7SUJ
 
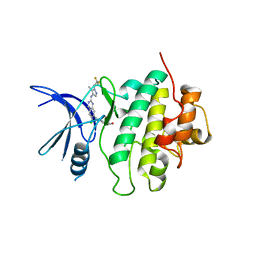 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 24 | | Descriptor: | (3R,4R)-4-{4-[6-chloro-2-({1-[(1R)-2,2-difluorocyclopropyl]-5-methyl-1H-pyrazol-4-yl}amino)quinazolin-7-yl]piperidin-1-yl}-4-methyloxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
6NJA
 
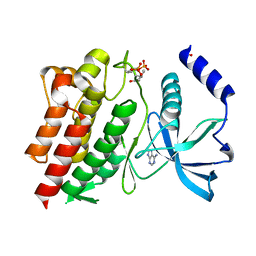 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7TE8
 
 | | CA14-CBD-DB21 ternary complex | | Descriptor: | CA14, DB21, cannabidiol | | Authors: | Cao, S, Zheng, N. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Defining molecular glues with a dual-nanobody cannabidiol sensor.
Nat Commun, 13, 2022
|
|
8J4V
 
 | |
9KCT
 
 | | Crystal structure of Tagatose 4-epimerase from Thermoprotei archaeon | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, J, Luo, G, Huang, Z, Ni, D, Zhu, Y, Xu, W, Zhang, W, Mu, W. | | Deposit date: | 2024-11-02 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Tagatose 4-epimerase from Thermoprotei archaeon
To Be Published
|
|
8XGJ
 
 | | Human Cx36/GJD2 gap junction channel in complex with mefloquine. | | Descriptor: | (11R,12S)- Mefloquine, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
6NQW
 
 | | Flagellar protein FcpA from Leptospira biflexa - hexagonal form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Flagellar coiling protein A, GLYCEROL, ... | | Authors: | San Martin, F, Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
6CUQ
 
 | |
8JAG
 
 | |
8OUV
 
 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
7PLQ
 
 | | Crystal structure of the PARP domain of wheat SRO1 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Wirthmueller, L, Loll, B. | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The superior salinity tolerance of bread wheat cultivar Shanrong No. 3 is unlikely to be caused by elevated Ta-sro1 poly-(ADP-ribose) polymerase activity.
Plant Cell, 34, 2022
|
|
8ZTO
 
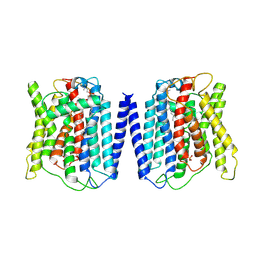 | | Cryo-EM structure of the human XPR1 | | Descriptor: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate | | Authors: | She, J, Chen, L. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure and function of human XPR1 in phosphate export.
Nat Commun, 16, 2025
|
|
5H80
 
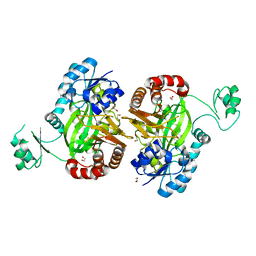 | | Biotin Carboxylase domain of single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
8T3X
 
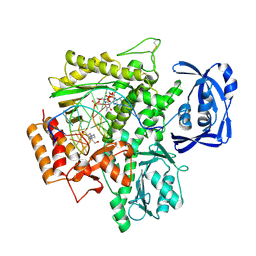 | | TNA polymerase, closed ternary | | Descriptor: | 10-92, TNA polymerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Maola, V, Chaput, J, Chim, N. | | Deposit date: | 2023-06-07 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Directed evolution of a highly efficient TNA polymerase achieved by homologous recombination
Nat Catal, 2024
|
|
6R0U
 
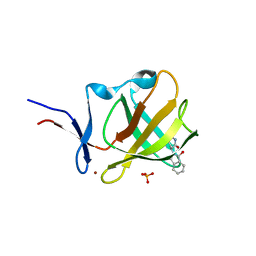 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5a and hydrolysis product | | Descriptor: | 3-azanyl-2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]benzoic acid, 4-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0S
 
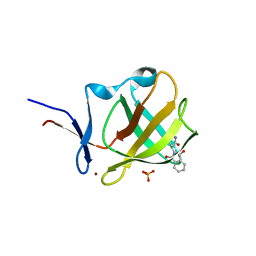 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 4a and hydrolysis product | | Descriptor: | 2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]-4-nitro-isoindole-1,3-dione, 2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]-6-nitro-benzoic acid, CEREBLON ISOFORM 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
5HBB
 
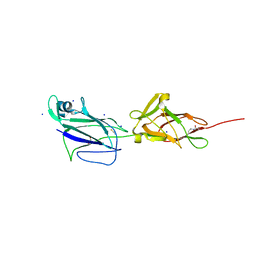 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6R11
 
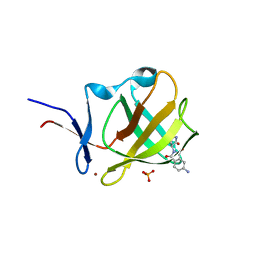 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5b | | Descriptor: | 5-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, Cereblon isoform 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
