6IAG
 
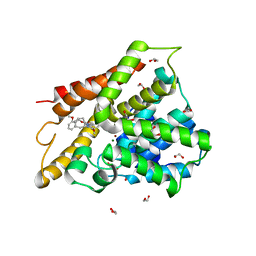 | |
6IBF
 
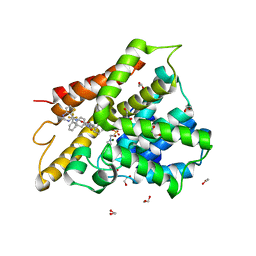 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-417 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-4-[4-methoxy-3-[[2-(trifluoromethyl)phenyl]methoxy]phenyl]-2-(1-thieno[3,2-d]pyrimidin-4-ylpiperidin-4-yl)-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-11-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-417
To be published
|
|
7Y74
 
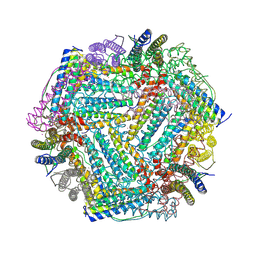 | |
7X8X
 
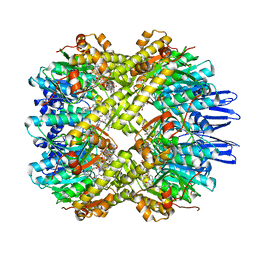 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
7X0S
 
 | | Human TRiC-tubulin-S3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7XNY
 
 | |
6I7V
 
 | | Ribosomal protein paralogs bL31 and bL36 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Pulk, A, Cate, J.H.D, Remme, J, Lilleorg, S, Reier, K, Peil, L, Liiv, A, Tammsalu, T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacterial ribosome heterogeneity: Changes in ribosomal protein composition during transition into stationary growth phase.
Biochimie, 156, 2018
|
|
6U5B
 
 | | CryoEM Structure of Pyocin R2 - precontracted - baseplate | | Descriptor: | Glue PA0627, Ripcord PA0626, Sheath Initiator PA0617, ... | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6I9R
 
 | | Large subunit of the human mitochondrial ribosome in complex with Virginiamycin M and Quinupristin | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Modelska, A, Aibara, S, Amunts, A. | | Deposit date: | 2018-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth.
Cell Rep, 35, 2021
|
|
7XJ6
 
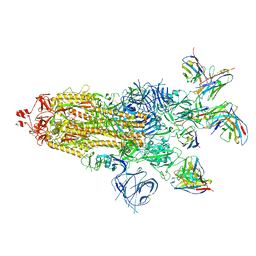 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
6IU9
 
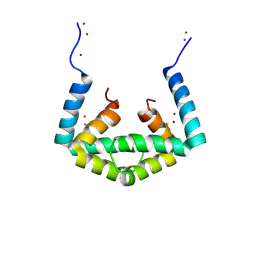 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6U0S
 
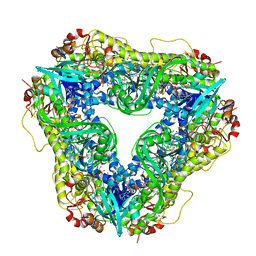 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
6U2J
 
 | | EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
4YA0
 
 | |
6IF5
 
 | | Crystal structure of monkey TLR7 in complex with 2',3'-cGMP (Guanosine 2',3'-cyclic phosphate) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5W4L
 
 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
4YCO
 
 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNAPhe | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VVC
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-(2-(((2-Amino-4-methylquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-{[(2-amino-4-methylquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4YEE
 
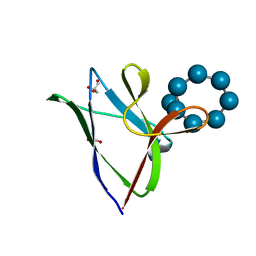 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
5W6R
 
 | |
6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
6U9V
 
 | |
