5EEU
 
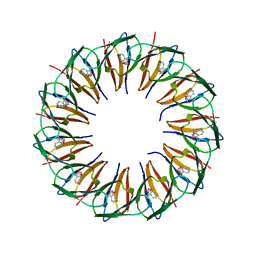 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 1.31 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5TPV
 
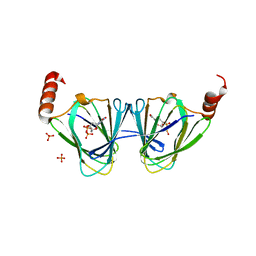 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
1OVH
 
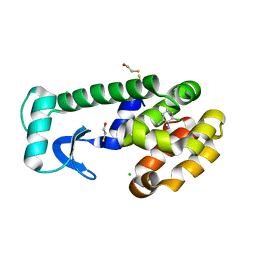 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Chloro-6-Methyl-Aniline | | Descriptor: | 2-CHLORO-6-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
3KOJ
 
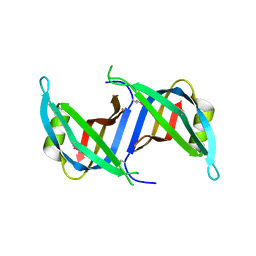 | | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp. Northeast Structural Genomics Consortium Target SnR59a. | | Descriptor: | uncharacterized protein ycf41 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Wang, H, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp.
To be Published
|
|
3KX2
 
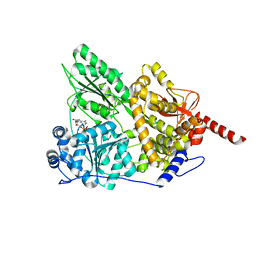 | |
3KOO
 
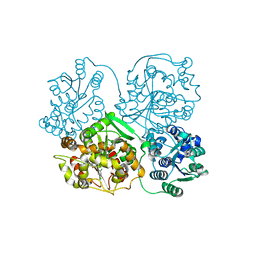 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | Epoxide hydrolase 2, N-(2,4-dichlorobenzyl)-4-(pyrimidin-2-yloxy)piperidine-1-carboxamide | | Authors: | Farrow, N.A. | | Deposit date: | 2009-11-13 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Optimization of piperidyl-ureas as inhibitors of soluble epoxide hydrolase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
2OSC
 
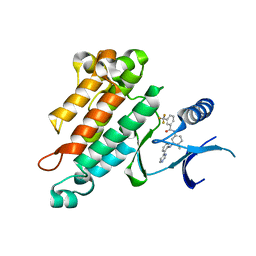 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1OWY
 
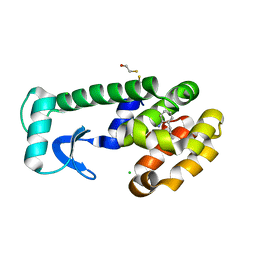 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Propyl-Aniline | | Descriptor: | 2-PROPYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
3KZ1
 
 | | Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Chen, Z, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2009-12-07 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.
J.Biol.Chem., 285, 2010
|
|
2OPX
 
 | |
3C6N
 
 | | Small molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling | | Descriptor: | (2S)-8-[(tert-butoxycarbonyl)amino]-2-(1H-indol-3-yl)octanoic acid, INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, ... | | Authors: | Tan, X, Zheng, N, Hayashi, K. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3KZS
 
 | |
5FAX
 
 | |
3L1R
 
 | |
3CA6
 
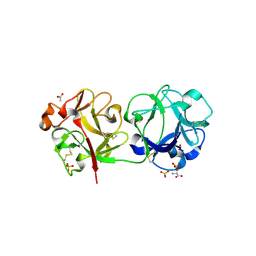 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to Tn antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
5UHK
 
 | |
3KSI
 
 | |
1P1O
 
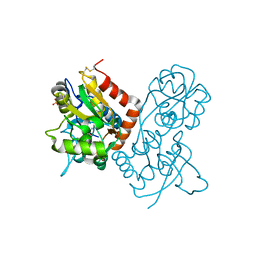 | | Crystal structure of the GluR2 ligand-binding core (S1S2J) mutant L650T in complex with quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Armstrong, N, Mayer, M.L, Gouaux, E. | | Deposit date: | 2003-04-13 | | Release date: | 2003-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic
adjustment of agonist-induced conformational changes.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2PHM
 
 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
5FZ0
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide (N08137b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Pearce, N, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N11213A
To be Published
|
|
3BNK
 
 | |
5FKE
 
 | |
5TWQ
 
 | | Post-catalytic nicked complex of human Polymerase Mu with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5FLJ
 
 | | enzyme-substrate-dioxygen complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, NICKEL (II) ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
