5FZ6
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment N05859b (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-CARBOXYPIPERIDINE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment N05859B (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
5FYI
 
 | | Crystal structure of human JMJD2A in complex with pyruvate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Szykowska, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal Structure of Human Jmjd2A in Complex with Pyruvate
To be Published
|
|
5FYH
 
 | | Crystal structure of human JMJD2A in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Szykowska, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal Structure of Human Jmjd2A in Complex with Fumarate
To be Published
|
|
3IQQ
 
 | | X-ray structure of bovine TRTK12-Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TRTK12 peptide, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-08-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Effects of CapZ Peptide (TRTK-12) Binding to S100B-Ca(2+) as Examined by NMR and X-ray Crystallography
J.Mol.Biol., 396, 2010
|
|
5FY5
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Fumarate
To be Published
|
|
2NBQ
 
 | |
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
5FYC
 
 | | Crystal structure of human JMJD2A in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Szykowska, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal Structure of Human Jmjd2A in Complex with Succinate
To be Published
|
|
3IRL
 
 | | Solution Structure of Heparin dp36 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
5FY4
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Succinate
To be Published
|
|
5FY9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with pyruvate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Pyruvate
To be Published
|
|
1QW4
 
 | | Crystal Structure of Murine Inducible Nitric Oxide Synthase Oxygenase Domain in complex with N-omega-propyl-L-arginine. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-PROPYL-L-ARGININE, Nitric oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-08-31 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
5UGX
 
 | |
3ISE
 
 | | Structure of mineralized Bfrb (double soak) from Pseudomonas aeruginosa to 2.8A Resolution | | Descriptor: | Bacterioferritin, FE (III) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3AB2
 
 | | Crystal structure of aspartate kinase from Corynebacterium glutamicum in complex with threonine | | Descriptor: | Aspartokinase, THREONINE | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
5FUN
 
 | | Crystal structure of human JARID1B in complex with GSK467 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(1-benzyl-1H-pyrazol-4-yl)oxy]pyrido[3,4-d]pyrimidin-4(3H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FVK
 
 | | Crystal structure of Vps4-Vfa1 complex from S.cerevisiae at 1.66 A resolution. | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4, VPS4-ASSOCIATED PROTEIN 1 | | Authors: | Kojima, R, Obita, T, Onoue, K, Mizuguchi, M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Structural Fine-Tuning of Mit Interacting Motif 2 (Mim2) and Allosteric Regulation of Escrt-III by Vps4 in Yeast.
J.Mol.Biol., 428, 2016
|
|
3A5M
 
 | | Crystal Structure of a Dictyostelium P109I Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
1QMT
 
 | |
2N5X
 
 | |
5FJH
 
 | |
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3IUD
 
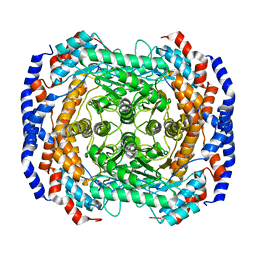 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
2ZX8
 
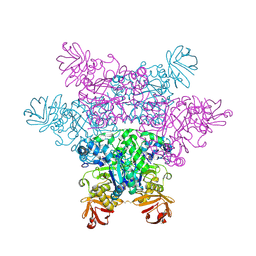 | | alpha-L-fucosidase complexed with inhibitor, F10-2C-O | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1-benzofuran-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
