5DSD
 
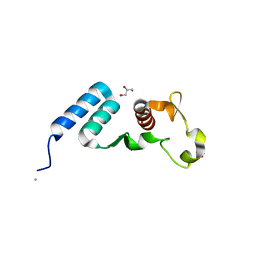 | | The crystal structure of the C-terminal domain of Ebola (Bundibugyo) nucleoprotein | | Descriptor: | CHLORIDE ION, GLYCEROL, Nucleoprotein | | Authors: | Baker, L, Handing, K.B, Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3H11
 
 | |
4IDA
 
 | |
2G4E
 
 | |
5DTK
 
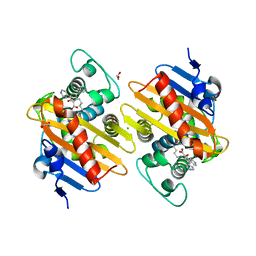 | | Fragments bound to the OXA-48 beta-lactamase: Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di(pyridin-4-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Christopeit, T, Leiros, H.-K.S. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.60000241 Å) | | Cite: | Screening and Design of Inhibitor Scaffolds for the Antibiotic Resistance Oxacillinase-48 (OXA-48) through Surface Plasmon Resonance Screening.
J.Med.Chem., 59, 2016
|
|
2G5G
 
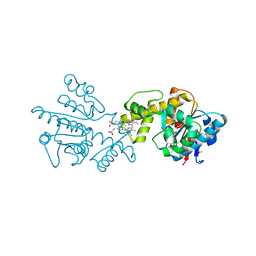 | |
1J1C
 
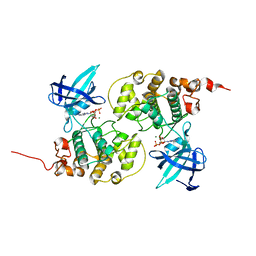 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4IJ4
 
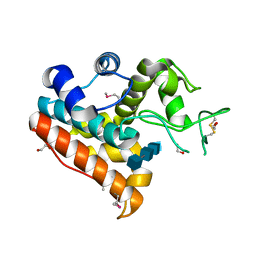 | | Crystal Structure of a Family GH19 chitinase from Bryum coronatum in complex with (GlcNAc)4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-12-21 | | Release date: | 2014-03-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
6CBK
 
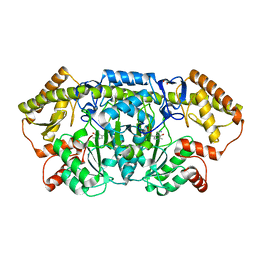 | | X-ray structure of NeoB from Streptomyces fradiae in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Neamine transaminase NeoN, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
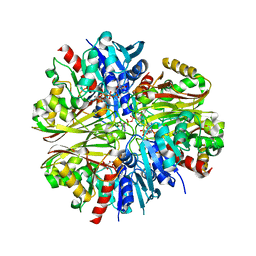
4K9D
 
 | |
3H7G
 
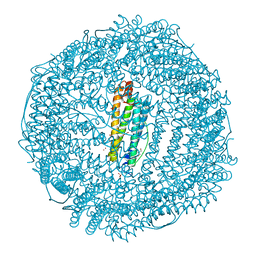 | | Apo-FR with AU ions | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Abe, M, Ueno, T, Abe, S, Suzuki, M, Goto, T, Toda, Y, Akita, T, Yamada, Y, Watanabe, Y. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Preparation and catalytic reaction of Au/Pd bimetallic nanoparticles in apo-ferritin
Chem.Commun.(Camb.), 32, 2009
|
|
2YOL
 
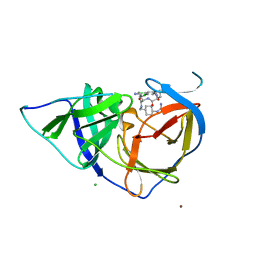 | | West Nile Virus NS2B-NS3 protease in complex with 3,4- dichlorophenylacetyl-Lys-Lys-GCMA | | Descriptor: | (S)-6-amino-N-((S)-6-amino-1-(((1r,4S)-4-guanidinocyclohexyl)methylamino)-1-oxohexan-2-yl)-2-(2-(3,4-dichlorophenyl)acetamido)hexanamide, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Hammamy, M.Z, Haase, C, Hammami, M, Hilgenfeld, R, Steinmetzer, T. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Development and Characterization of New Peptidomimetic Inhibitors of the West Nile Virus Ns2B-Ns3 Protease.
Chemmedchem, 8, 2013
|
|
5DVH
 
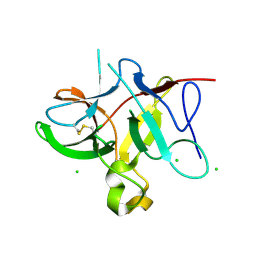 | |
3H8O
 
 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*(MA7)P*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*TP*AP*TP*AP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, Q. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
6CB7
 
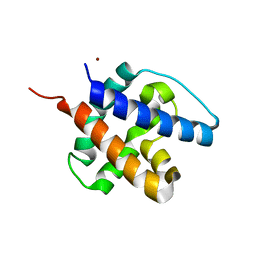 | |
2GI9
 
 | |
6CBL
 
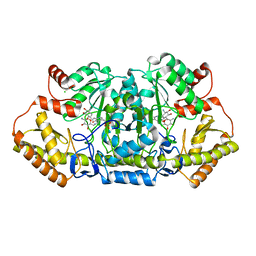 | | x-ray structure of NeoB from Streptomyces fradiae in complex with neamine as an external aldimine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, CHLORIDE ION, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CF3
 
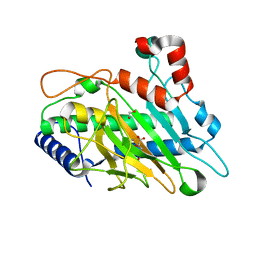 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
5E8I
 
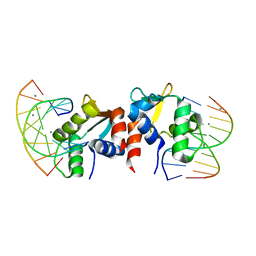 | |
1J8T
 
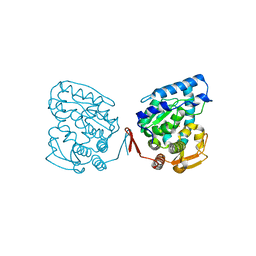 | | Catalytic Domain of Human Phenylalanine Hydroxylase Fe(II) | | Descriptor: | FE (II) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin.
J.Mol.Biol., 314, 2001
|
|
2YW4
 
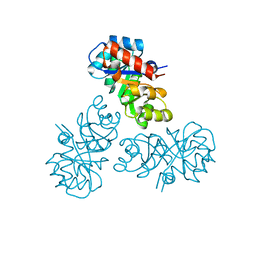 | |
6CCM
 
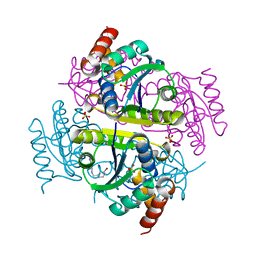 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
 | |
2G01
 
 | | Pyrazoloquinolones as Novel, Selective JNK1 inhibitors | | Descriptor: | 6-CHLORO-9-HYDROXY-1,3-DIMETHYL-1,9-DIHYDRO-4H-PYRAZOLO[3,4-B]QUINOLIN-4-ONE, C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-02-10 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6CD2
 
 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
