4ZA4
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
1KVT
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
4JZZ
 
 | |
6BFZ
 
 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
5D3X
 
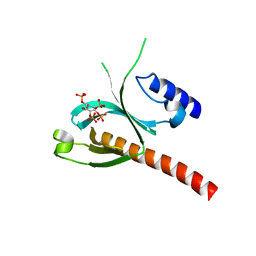 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
3FJF
 
 | |
4ZE6
 
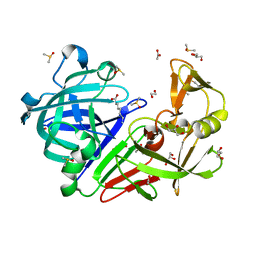 | | Endothiapepsin in complex with fragment B39 | | Descriptor: | 1,2-ETHANEDIOL, 7-aminoheptanoic acid, ACETATE ION, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-04-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5CU2
 
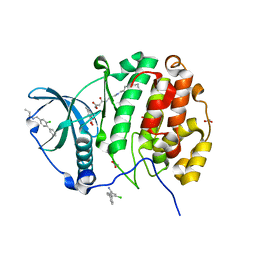 | | Crystal structure of CK2alpha with 2-hydroxy-5-methylbenzoic acid and (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoat bound | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
4JYW
 
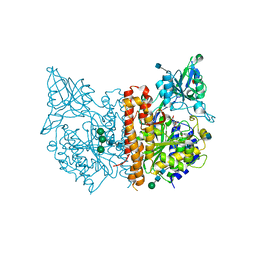 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with CTT1057 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A high affinity 18F-labeled phosphoramidate peptidomimetic inhibitor as a PSMA-targeted PET imaging agent for prostate cancer
To be Published
|
|
4YXU
 
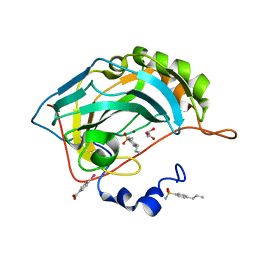 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (4). | | Descriptor: | 4-propylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
2V62
 
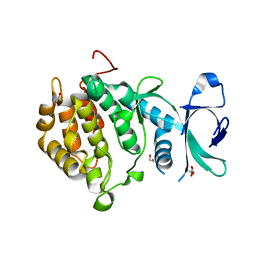 | | Structure of vaccinia-related kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SERINE/THREONINE-PROTEIN KINASE VRK2, ... | | Authors: | Bunkoczi, G, Eswaran, J, Cooper, C, Fedorov, O, Keates, T, Rellos, P, Salah, E, Savitsky, P, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Pseudokinase Vrk3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site.
Structure, 17, 2009
|
|
5CX1
 
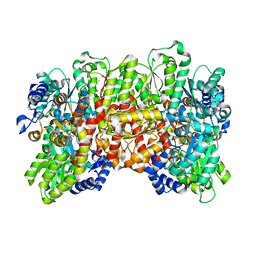 | | Nitrogenase molybdenum-iron protein beta-K400E mutant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Luca, M.A, Tezcan, F.A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7476 Å) | | Cite: | Evidence for Functionally Relevant Encounter Complexes in Nitrogenase Catalysis.
J.Am.Chem.Soc., 137, 2015
|
|
2V5F
 
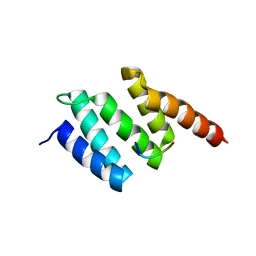 | | Crystal structure of wild type peptide-binding domain of human type I collagen prolyl 4-hydroxylase. | | Descriptor: | HEXA-HISTIDINE PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Pekkala, M, Hieta, R, Kivirikko, K, Myllyharju, J, Wierenga, R. | | Deposit date: | 2008-10-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Wild Type Peptide-Binding Domain of Human Type I Collagen Prolyl 4- Hydroxylase.
To be Published
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1KEK
 
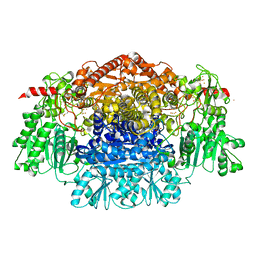 | | Crystal Structure of the Free Radical Intermediate of Pyruvate:Ferredoxin Oxidoreductase | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Vernede, X, Guigliarelli, B, Charon, M.-H, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2001-11-16 | | Release date: | 2001-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase.
Science, 294, 2001
|
|
4YZT
 
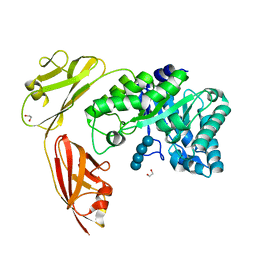 | | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, Cellulose hydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Sci Rep, 6, 2016
|
|
4Z0B
 
 | | Crystal Structure of the Fab Fragment of Anti-ofloxacin Antibody and Exploration Its Receptor Binding Site | | Descriptor: | PHOSPHATE ION, antibody heavy chain, antibody light chain | | Authors: | He, K, Du, X, Sheng, W, Zhou, X, Wang, J, Wang, S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-ofloxacin Antibody and Exploration of Its Specific Binding.
J.Agric.Food Chem., 64, 2016
|
|
2VJ4
 
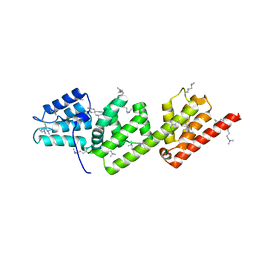 | | Methylated Shigella flexneri MxiC | | Descriptor: | PROTEIN MXIC | | Authors: | Deane, J.E, Roversi, P, King, C, Johnson, S, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the Shigella Flexneri Type 3 Secretion System Protein Mxic Reveal Conformational Variability Amongst Homologues.
J.Mol.Biol., 377, 2008
|
|
4K26
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 -HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Loenze, P, Schimanski-Breves, S, Vonderheyden, C, Engel, C.K. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4Z1N
 
 | | Carbonic anhydrase inhibitors: Design and synthesis of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms (hCA VII, hCA IX, and hCA XIV) | | Descriptor: | 4-[(6,7-dimethoxy-3,4-dihydro-1H-isoquinolin-2-yl)carbonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Pospisilova, K, Rezacova, P, Pachl, P. | | Deposit date: | 2015-03-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbonic anhydrase inhibitors: Design, synthesis and structural characterization of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms.
Eur.J.Med.Chem., 102, 2015
|
|
2FU9
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (mp2 inhibitor complex) | | Descriptor: | GLYCEROL, Metallo-beta-lactamase L1, N-[(BENZYLOXY)CARBONYL]-L-CYSTEINYLGLYCINE, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-26 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
5D29
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU241 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{S})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-08-05 | | Release date: | 2016-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
5D3K
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
1KI6
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | Descriptor: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
