2CB2
 
 | | Sulfur Oxygenase Reductase from Acidianus Ambivalens | | Descriptor: | FE (III) ION, SULFUR OXYGENASE REDUCTASE | | Authors: | Urich, T, Gomes, C.M, Kletzin, A, Frazao, C. | | Deposit date: | 2005-12-28 | | Release date: | 2006-02-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Structure of a Self-Compartmentalizing Sulfur Cycle Metalloenzyme
Science, 311, 2006
|
|
5Z2P
 
 | | ThDP-Mn2+ complex of R413K variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
6XF8
 
 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
4YYT
 
 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (5). | | Descriptor: | 4-(2-hydroxyethyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
5YUQ
 
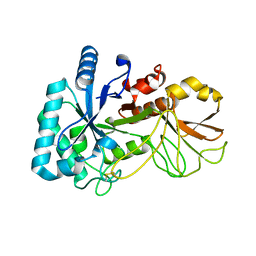 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | Descriptor: | Chintase | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
5DTD
 
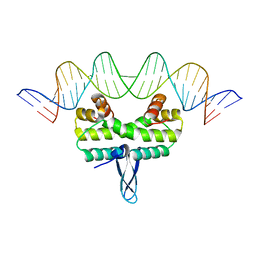 | |
3DUF
 
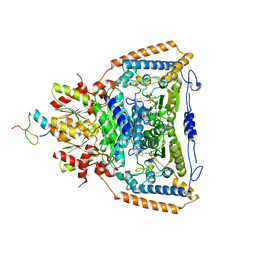 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
6XHH
 
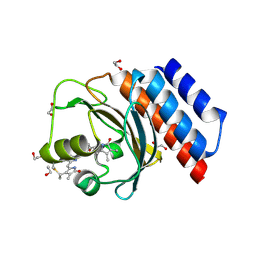 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
4YZP
 
 | |
6Y15
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E_Q126K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
1JUD
 
 | | L-2-HALOACID DEHALOGENASE | | Descriptor: | L-2-HALOACID DEHALOGENASE | | Authors: | Hisano, T, Hata, Y, Fujii, T, Liu, J.-Q, Kurihara, T, Esaki, N, Soda, K. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold.
J.Biol.Chem., 271, 1996
|
|
6Y1B
 
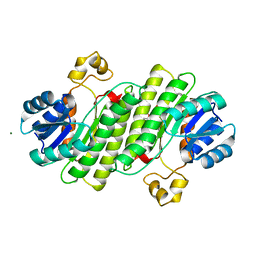 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
3DWL
 
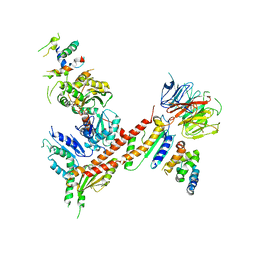 | |
3DXU
 
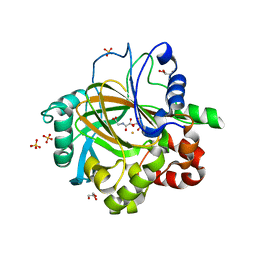 | |
2CBD
 
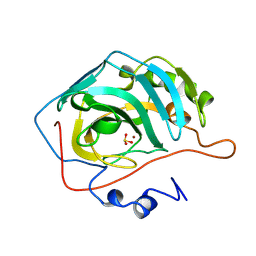 | | STRUCTURE OF NATIVE AND APO CARBONIC ANHYDRASE II AND SOME OF ITS ANION-LIGAND COMPLEXES | | Descriptor: | CARBONIC ANHYDRASE II, SULFITE ION, ZINC ION | | Authors: | Hakansson, K, Carlsson, M, Svensson, L.A, Liljas, A. | | Deposit date: | 1992-06-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes.
J.Mol.Biol., 227, 1992
|
|
4Z1K
 
 | | Carbonic anhydrase inhibitors: Design and synthesis of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms (hCA VII, hCA IX, and hCA XIV) | | Descriptor: | 4-[(6,7-dihydroxy-3,4-dihydroisoquinolin-2(1H)-yl)carbonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Pospisilova, K, Rezacova, P, Pachl, P. | | Deposit date: | 2015-03-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic anhydrase inhibitors: Design, synthesis and structural characterization of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms.
Eur.J.Med.Chem., 102, 2015
|
|
4Z1R
 
 | | Crystal structure of collagen-like peptide at 1.27 Angstrom resolution | | Descriptor: | Collagen-like peptide | | Authors: | Plonska-Brzezinska, M.E, Czyrko, J, Brus, D.M, Imierska, M, Brzezinski, K. | | Deposit date: | 2015-03-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Triple helical collagen-like peptide interactions with selected polyphenolic compounds.
Rsc Adv, 5, 2015
|
|
1K2O
 
 | | Cytochrome P450Cam with Bound BIS(2,2'-BIPYRIDINE)-(5-METHYL-2-2'-BIPYRIDINE)-C2-ADAMANTANE RUTHENIUM (II) | | Descriptor: | CACODYLATE ION, Cytochrome P450CAM, DELTA-BIS(2,2'-BIPYRIDINE)-(5-METHYL-2-2'-BIPYRIDINE)-C2-ADAMANTANE RUTHENIUM (II), ... | | Authors: | Dunn, A.R, Dmochowski, I.J, Bilwes, A.M, Gray, H.B, Crane, B.R. | | Deposit date: | 2001-09-28 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the open state of cytochrome P450cam with ruthenium-linker substrates.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6Y36
 
 | | CCAAT-binding complex from Aspergillus fumigatus with cccA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
3DYS
 
 | | human phosphodiestrase-5'GMP complex (EP), produced by soaking with 20mM cGMP+20 mM MnCl2+20 mM MgCl2 for 2 hours, and flash-cooled to liquid nitrogen temperature when substrate was still abudant. | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, FORMIC ACID, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4Z2T
 
 | |
2VNL
 
 | | MUTANT Y108Wdel OF THE HEADBINDING DOMAIN OF PHAGE P22 TAILSPIKE C- TERMINally fused to ISOLEUCINE ZIPPER pIIGCN4 (chimera II) | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, PIIGCN4, GLYCEROL | | Authors: | Mueller, J.J, Seul, A, Mueller, G, Seckler, R, Heinemann, U. | | Deposit date: | 2008-02-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Z26
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
4JD1
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | FOSFOMYCIN, Metallothiol transferase FosB 2, TRIETHYLENE GLYCOL, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Sharma, S.V, Hamilton, C.J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
