2VSR
 
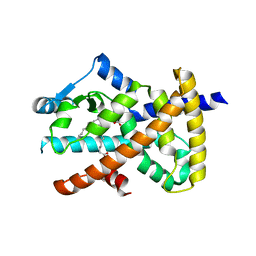 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
1KW0
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase (Fe(II)) in Complex with Tetrahydrobiopterin and Thienylalanine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, BETA(2-THIENYL)ALANINE, FE (II) ION, ... | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Ternary Complex of the Catalytic
Domain of Human Phenylalanine Hydroxylase with Tetrahydrobiopterin
and 3-(2-thienyl)-L-alanine, and its Implications for the Mechanism
of Catalysis and Substrate Activation
J.Mol.Biol., 320, 2002
|
|
4Y5C
 
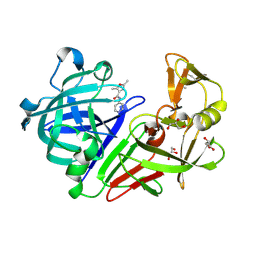 | | Endothiapepsin in complex with fragment 267 | | Descriptor: | 2,5-dimethyl-N-(pyridin-4-yl)furan-3-carboxamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
2E03
 
 | |
4Y5M
 
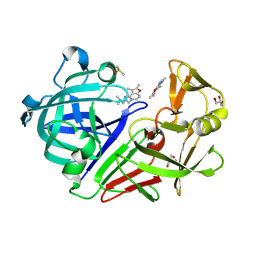 | | Endothiapepsin in complex with fragment 328 | | Descriptor: | 3-[(4E)-4-imino-5,6-dimethylfuro[2,3-d]pyrimidin-3(4H)-yl]-N,N-dimethylpropan-1-amine, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Fu, K, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4J0Q
 
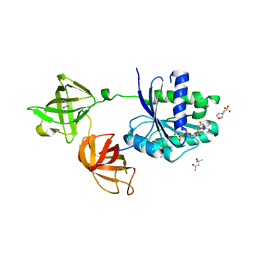 | | Crystal structure of Pseudomonas putida elongation factor Tu (EF-Tu) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Elongation factor Tu-A, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6AD4
 
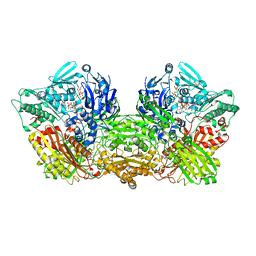 | | Rat Xanthine oxidoreductase, D428A variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
3FXS
 
 | |
6AHR
 
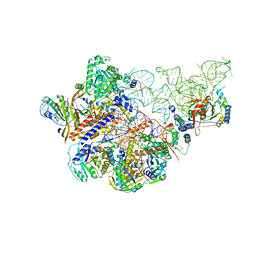 | | Cryo-EM structure of human Ribonuclease P | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
4J3J
 
 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
2VQG
 
 | | Crystal structure of PorB from Corynebacterium glutamicum (crystal form I) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 3,6,9,12,15,18,21,24,27-nonaoxaheptatriacontan-1-ol, ACETATE ION, ... | | Authors: | Ziegler, K, Benz, R, Schulz, G.E. | | Deposit date: | 2008-03-15 | | Release date: | 2008-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A Putative Alpha-Helical Porin from Corynebacterium Glutamicum.
J.Mol.Biol., 379, 2008
|
|
6AJU
 
 | | Rat Xanthine oxidoreductase | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
2VS4
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
6ALQ
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
3G0D
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 2 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
4YHV
 
 | | Yeast Prp3 C-terminal fragment 325-469 | | Descriptor: | ACETIC ACID, U4/U6 small nuclear ribonucleoprotein PRP3 | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
6ALV
 
 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6X3M
 
 | | Crystal structure of full-length Streptococcal bacteriophage hyaluronidase in complex with unsaturated hyaluronan octa-saccharides | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronoglucosaminidase | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
5C5W
 
 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2VXO
 
 | | Human GMP synthetase in complex with XMP | | Descriptor: | GMP SYNTHASE [GLUTAMINE-HYDROLYZING], SULFATE ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Welin, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-07-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity and Oligomerization of Human Gmp Synthetase
J.Mol.Biol., 425, 2013
|
|
6AM0
 
 | |
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
5C9J
 
 | | Human CD1c with ligands in A' and F' channel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, LAURIC ACID, ... | | Authors: | Tews, I, Machelett, M.M, Mansour, S, Gadola, S.D. | | Deposit date: | 2015-06-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y3N
 
 | | Endothiapepsin in complex with fragment 273 | | Descriptor: | 2-[(1S)-2-acetyl-1,2-dihydroisoquinolin-1-yl]-N,N-dimethylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4IFH
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | Descriptor: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | Authors: | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
