4XOT
 
 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
5C6B
 
 | |
6WUB
 
 | |
4XN3
 
 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
3FA4
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, triclinic crystal form | | Descriptor: | 2,3-dimethylmalate lyase, MAGNESIUM ION | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
6AK1
 
 | | Crystal structure of DmoA from Hyphomicrobium sulfonivorans | | Descriptor: | Dimethyl-sulfide monooxygenase | | Authors: | Cao, H.Y, Wang, P, Peng, M, Li, C.Y. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
6WZC
 
 | |
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
4HPD
 
 | |
3FBI
 
 | | Structure of the Mediator submodule Med7N/31 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | Authors: | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
1LGQ
 
 | | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein | | Descriptor: | cell cycle checkpoint protein CHFR | | Authors: | Stavridi, E.S, Huyen, Y, Loreto, I.R, Scolnick, D.M, Halazonetis, T.D, Pavletich, N.P, Jeffrey, P.D. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate.
Structure, 10, 2002
|
|
6WYF
 
 | |
4HWN
 
 | | Crystal structure of the second Ig-C2 domain of human Fc-receptor like A (FCRLA), Isoform 9 [NYSGRC-005836] | | Descriptor: | Fc receptor-like A | | Authors: | Kumar, P.R, Ahmed, M, Bhosle, R, Calarese, D, Celikigil, A, Chan, M.K, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of the second Ig-C2 domain of the human Fc-receptor like A, Isoform 9 [NYSGRC-005836]
to be published
|
|
4XON
 
 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
6X0R
 
 | |
3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
5C1B
 
 | |
5C0B
 
 | | 1E6 TCR in complex with HLA-A02 carrying RQFGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
6AK3
 
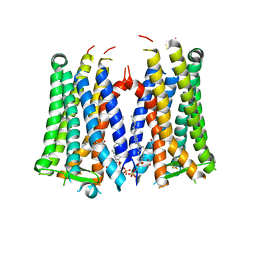 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | Authors: | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-08-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
4XQB
 
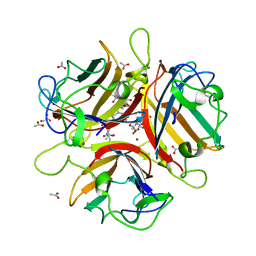 | | CRYSTAL STRUCTURE OF AD37 FIBER KNOB IN COMPLEX WITH TRIVALENT SIALIC ACID INHIBITOR ME0461 | | Descriptor: | 2-(1-{2-[bis(2-{4-[2-({(6R)-5-(acetylamino)-3,5-dideoxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-beta-L-threo-hex-2-ulopyranonosyl}oxy)ethyl]-1H-1,2,3-triazol-1-yl}ethyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)ethyl (6R)-5-(acetylamino)-3,5-dideoxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-beta-L-threo-hex-2-ulopyranosidonic acid, ACETATE ION, Fiber, ... | | Authors: | Stehle, T, Liaci, A.M. | | Deposit date: | 2015-01-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
3FAA
 
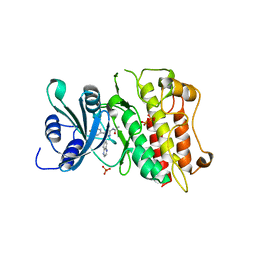 | |
2WIE
 
 | | High-resolution structure of the rotor ring from a proton dependent ATP synthase | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | High-Resolution Structure of the Rotor Ring of a Proton-Dependent ATP Synthase.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4XR6
 
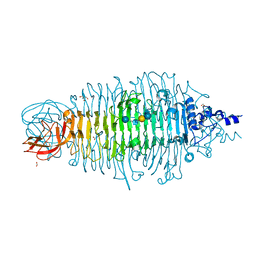 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
3FB7
 
 | | Open KcsA potassium channel in the presence of Rb+ ion | | Descriptor: | RUBIDIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Pan, A.C, Gagnon, D.H, Cordero-Morales, J.F, Chakrapani, S, Roux, B, Perozo, E. | | Deposit date: | 2008-11-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open KcsA potassium channel in the presence of Rb+ ion
TO BE PUBLISHED
|
|
3FBX
 
 | | Crystal structure of the lysosomal 66.3 kDa protein from mouse solved by S-SAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Mueller, U, Ficner, R. | | Deposit date: | 2008-11-20 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo sulfur SAD phasing of the lysosomal 66.3 kDa protein from mouse
Acta Crystallogr.,Sect.D, 65, 2009
|
|
