4BXX
 
 | | Arrested RNA polymerase II-Bye1 complex | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*AP*CP*CP*TP*GP *GP*TP*GP* BRUP*TP*GP*CP*TP*CP*TP*AP*AP*DC)-3', 5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP)-3', 5'-D(*GP*AP*GP*GP*TP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6FYX
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
2PMZ
 
 | | Archaeal RNA polymerase from Sulfolobus solfataricus | | Descriptor: | DNA-directed RNA polymerase subunit A, DNA-directed RNA polymerase subunit A", DNA-directed RNA polymerase subunit B, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea
Nature, 451, 2008
|
|
2R7Z
 
 | | Cisplatin lesion containing RNA polymerase II elongation complex | | Descriptor: | 5'-D(*CP*AP*AP*GP*TP*AP*G)-3', 5'-D(*TP*AP*CP*TP*TP*GUP*CP*CP*CP*TP*CP*CP*TP*CP*AP*T)-3', 5'-R(*UP*UP*UP*GP*AP*GP*GP*AP*GP*G)-3', ... | | Authors: | Damsma, G.E, Alt, A, Brueckner, F, Carell, T, Cramer, P. | | Deposit date: | 2007-09-10 | | Release date: | 2007-11-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of transcriptional stalling at cisplatin-damaged DNA.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2R92
 
 | | Elongation complex of RNA polymerase II with artificial RdRP scaffold | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Lehmann, E, Brueckner, F, Cramer, P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
5U5Q
 
 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5C3E
 
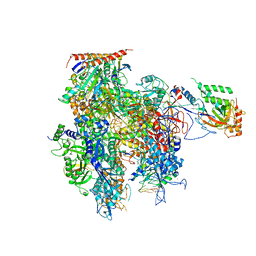 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C44
 
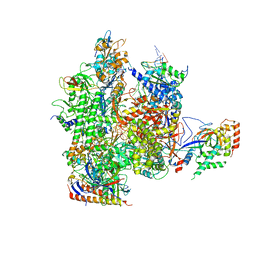 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5MQF
 
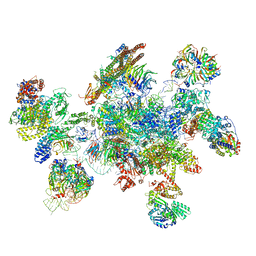 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5FLM
 
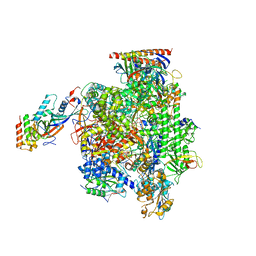 | | Structure of transcribing mammalian RNA polymerase II | | Descriptor: | DNA, DNA-RNA ELONGATION SCAFFOLD, DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Bernecky, C, Herzog, F, Baumeister, W, Plitzko, J.M, Cramer, P. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Transcribing Mammalian RNA Polymerase II
Nature, 529, 2016
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8H0V
 
 | | RNA polymerase II transcribing a chromatosome (type I) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8H0W
 
 | | RNA polymerase II transcribing a chromatosome (type II) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
5FYW
 
 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
8HE5
 
 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
5SVA
 
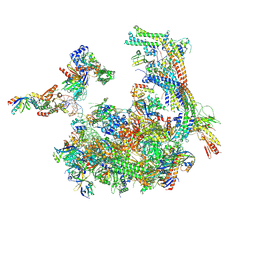 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | Descriptor: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | Authors: | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
5U0S
 
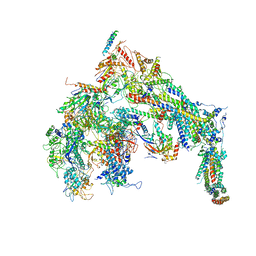 | | Cryo-EM structure of the Mediator-RNAPII complex | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
5FZ5
 
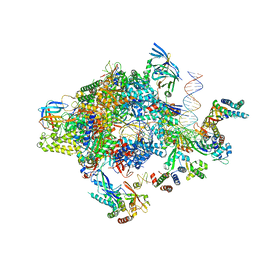 | | Transcription initiation complex structures elucidate DNA opening (CC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
5FMF
 
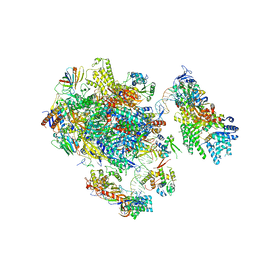 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VVR
 
 | |
5MS0
 
 | |
