1AK0
 
 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
1GYP
 
 | | CRYSTAL STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM LEISHMANIA MEXICANA: IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN AND A NEW POSITION FOR THE INORGANIC PHOSPHATE BINDING SITE | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Feil, I.K, Verlinde, C.L.M.J, Petra, P.H, Hol, W.G.J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Leishmania mexicana: implications for structure-based drug design and a new position for the inorganic phosphate binding site.
Biochemistry, 34, 1995
|
|
2YC1
 
 | | Crystal structure of the human derived single chain antibody fragment (scFv) 9004G in complex with Cn2 toxin from the scorpion Centruroides noxius Hoffmann | | Descriptor: | BETA-MAMMAL TOXIN CN2, GLYCEROL, SINGLE CHAIN ANTIBODY FRAGMENT 9004G | | Authors: | Canul-Tec, J.C, Riano-Umbarila, L, Rudino-Pinera, E, Becerril, B, Possani, L.D, Torres-Larios, A. | | Deposit date: | 2011-03-10 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Neutralization of the Major Toxic Component from the Scorpion Centruroides Noxius Hoffmann by a Human-Derived Single Chain Antibody Fragment.
J.Biol.Chem., 286, 2011
|
|
1GOW
 
 | |
3ZBY
 
 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1GRB
 
 | |
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3OUJ
 
 | | PHD2 with 2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Staker, B.L, Arakaki, T.L. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
158L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
167L
 
 | |
1HD7
 
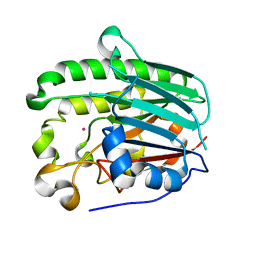 | | A Second Divalent Metal Ion in the Active Site of a New Crystal Form of Human Apurinic/Apyridinimic Endonuclease, Ape1, and its Implications for the Catalytic Mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-11-09 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
2YIC
 
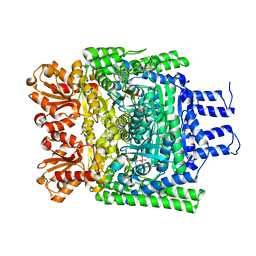 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
1USC
 
 | |
180L
 
 | |
4IHI
 
 | |
189L
 
 | |
1GT8
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1AKI
 
 | | THE STRUCTURE OF THE ORTHORHOMBIC FORM OF HEN EGG-WHITE LYSOZYME AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | LYSOZYME | | Authors: | Carter, D, He, J, Ruble, J.R, Wright, B. | | Deposit date: | 1997-05-19 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1LOI
 
 | | N-TERMINAL SPLICE REGION OF RAT C-AMP PHOSPHODIESTERASE, NMR, 7 STRUCTURES | | Descriptor: | CYCLIC 3',5'-AMP SPECIFIC PHOSPHODIESTERASE RD1 | | Authors: | Smith, K.J, Scotland, G, Beattie, J, Trayer, I.P, Houslay, M.D. | | Deposit date: | 1996-05-21 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of the N-terminal splice region of the cyclic AMP-specific phosphodiesterase RD1 (RNPDE4A1) by 1H NMR and identification of the membrane association domain using chimeric constructs.
J.Biol.Chem., 271, 1996
|
|
195L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
3WY4
 
 | | Crystal structure of alpha-glucosidase mutant E271Q in complex with maltose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
