1ULS
 
 | | Crystal structure of tt0140 from Thermus thermophilus HB8 | | Descriptor: | putative 3-oxoacyl-acyl carrier protein reductase | | Authors: | Hisanaga, Y, Ago, H, Hamada, K, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tt0140 from Thermus thermophilus HB8
To be Published
|
|
3ATR
 
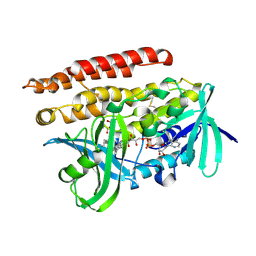 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius co-crystallized with its ligand | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, PYROPHOSPHATE, ... | | Authors: | Fujihashi, M, Sasaki, D, Iwata, Y, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
3P6P
 
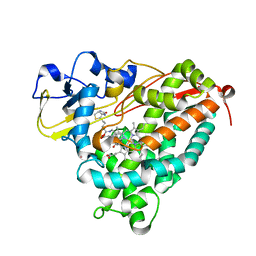 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio | | Descriptor: | (3S,5S,7S)-N-[6-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexyl]tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio
To be Published
|
|
1FWA
 
 | |
3WLN
 
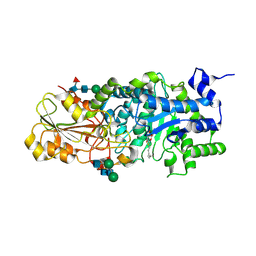 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
4IXL
 
 | | Crystal structure of endo-beta-1,4-xylanase from the alkaliphilic Bacillus sp. SN5 | | Descriptor: | SULFATE ION, xylanase | | Authors: | Bai, W.Q, Xue, Y.F, Huang, J.X, Zhou, C, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-01-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure Analysis of Alkaline xylanase from Alkaliphilic Bacillus sp. SN5 Implications for pH-dependent enzyme activity
To be Published
|
|
1B8S
 
 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES GLU361GLN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
4IXV
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 2d: {(5R)-5-amino-5-carboxy-5-[1-(4-chlorobenzyl)piperidin-4-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J14
 
 | | Crystal Structure of Human Cytochrome P450 CYP46A1 with Posaconazole Bound | | Descriptor: | Cholesterol 24-hydroxylase, GLYCEROL, POSACONAZOLE, ... | | Authors: | Stout, C.D, Mast, N, Pikuleva, I.A. | | Deposit date: | 2013-01-31 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antifungal Azoles: Structural Insights into Undesired Tight Binding to Cholesterol-Metabolizing CYP46A1.
Mol.Pharmacol., 84, 2013
|
|
1UOK
 
 | | CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE | | Descriptor: | OLIGO-1,6-GLUCOSIDASE | | Authors: | Watanabe, K, Hata, Y, Kizaki, H, Katsube, Y, Suzuki, Y. | | Deposit date: | 1998-07-28 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 A resolution: structural characterization of proline-substitution sites for protein thermostabilization.
J.Mol.Biol., 269, 1997
|
|
1GAY
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | MUTANT LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GEI
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
4J0Q
 
 | | Crystal structure of Pseudomonas putida elongation factor Tu (EF-Tu) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Elongation factor Tu-A, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1VBJ
 
 | |
1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
3WRK
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of P450cam intermedite
To be Published
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
3ON3
 
 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
1FZ6
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II SOAKED IN 1 M METHANOL | | Descriptor: | CALCIUM ION, FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase.
J.Am.Chem.Soc., 123, 2001
|
|
3AYQ
 
 | | Crystal structure of inhibitor bound lysozyme from Meretrix lusoria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-[(5S,6R)-5-hydroxy-6-(hydroxymethyl)-2-oxo-5,6-dihydro-2H-pyran-3-yl]acetamide, Lysozyme | | Authors: | Yoneda, K, Kuwano, Y, Usui, T, Ogata, M, Suzuki, A, Araki, T. | | Deposit date: | 2011-05-13 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of inhibitor bound lysozyme from Meretrix lusoria
To be Published
|
|
3WSV
 
 | |
1UR5
 
 | | Stabilization of a Tetrameric Malate Dehydrogenase by Introduction of a Disulfide Bridge at the Dimer/Dimer Interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of a Tetrameric Malate Dehydrogenase by Introduction of a Disulfide Bridge at the Dimer-Dimer Interface
J.Mol.Biol., 334, 2003
|
|
3WSW
 
 | |
1B31
 
 | |
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
