1EVJ
 
 | | CRYSTAL STRUCTURE OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE (GFOR) DELTA1-22 S64D | | Descriptor: | GLUCOSE-FRUCTOSE OXIDOREDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lott, J.S, Halbig, D, Baker, H.M, Hardman, M.J, Sprenger, G.A, Baker, E.N. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation.
J.Mol.Biol., 304, 2000
|
|
2CZT
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
1EJG
 
 | | CRAMBIN AT ULTRA-HIGH RESOLUTION: VALENCE ELECTRON DENSITY. | | Descriptor: | CRAMBIN (PRO22,SER22/LEU25,ILE25) | | Authors: | Jelsch, C, Teeter, M.M, Lamzin, V, Pichon-Lesme, V, Blessing, B, Lecomte, C. | | Deposit date: | 2000-03-02 | | Release date: | 2000-04-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.54 Å) | | Cite: | Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1IZ1
 
 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1ERX
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 COMPLEXED WITH NO | | Descriptor: | 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, CITRIC ACID, NITRIC OXIDE, ... | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-03 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial.
Nat.Struct.Biol., 7, 2000
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EX2
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS MAF PROTEIN | | Descriptor: | PHOSPHATE ION, PROTEIN MAF, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2CVQ
 
 | | Crystal structure of NAD(H)-dependent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
3V7K
 
 | |
1EAJ
 
 | |
1EA1
 
 | | Cytochrome P450 14 alpha-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, CYTOCHROME P450 51-LIKE RV0764C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Poulos, T.L, Waterman, M.R. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Cytochrome P450 14Alpha -Sterol Demethylase (Cyp51) from Mycobacterium Tuberculosis in Complex with Azole Inhibitors
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2D2N
 
 | | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi | | Descriptor: | Giant hemoglobin, A1(b) globin chain, A2(a5) globin chain, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2D7G
 
 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
1ETR
 
 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
2D7H
 
 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
2D7E
 
 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
1WRS
 
 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1F4D
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE C146S, L143C COVALENTLY MODIFIED AT C143 WITH N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL | | Descriptor: | GLYCEROL, N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1WRT
 
 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
3VNF
 
 | | Structure of the ebolavirus protein VP24 from Sudan | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
146D
 
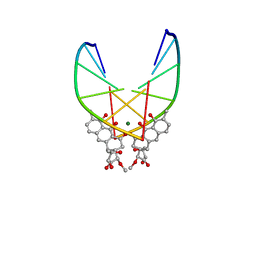 | | SOLUTION STRUCTURE OF THE MITHRAMYCIN DIMER-DNA COMPLEX | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*CP*GP*CP*GP*A)-3'), ... | | Authors: | Sastry, M, Patel, D.J. | | Deposit date: | 1993-11-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mithramycin dimer-DNA complex.
Biochemistry, 32, 1993
|
|
