1MHT
 
 | |
4JA7
 
 | | Rat PP5 co-crystallized with P5SA-2 | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
4JQV
 
 | | HLA-B*18:01 in complex with Epstein-Barr virus BZLF1-derived peptide (residues 173-180) | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Welland, A, Gras, S, Rossjohn, J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HLA Peptide Length Preferences Control CD8+ T Cell Responses.
J.Immunol., 191, 2013
|
|
1SAN
 
 | |
1WRI
 
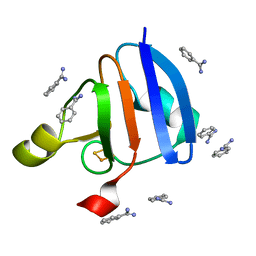 | | Crystal Structure of Ferredoxin isoform II from E. arvense | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin II | | Authors: | Kurisu, G, Nishiyama, D, Kusunoki, M, Fujikawa, S, Katoh, M, Hanke, G.T, Hase, T, Teshima, K. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structural basis of Equisetum arvense ferredoxin isoform II producing an alternative electron transfer with ferredoxin-NADP+ reductase.
J.Biol.Chem., 280, 2005
|
|
4JGH
 
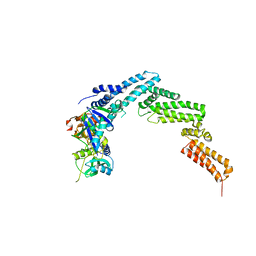 | | Structure of the SOCS2-Elongin BC complex bound to an N-terminal fragment of Cullin5 | | Descriptor: | Cullin-5, Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, ... | | Authors: | Kim, Y.K, Kwak, M.J, Ku, B, Suh, H.Y, Joo, K, Lee, J, Jung, J.U, Oh, B.H. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1RZF
 
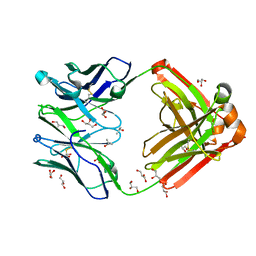 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4JTJ
 
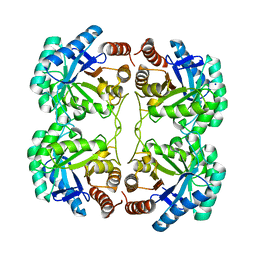 | | Crystal structure of R117K mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, SODIUM ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTL
 
 | | Crystal structure of F139G mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
1MUV
 
 | | Sheared A(anti)-A(anti) Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of 5'(rGGCAAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Schroeder, S.J, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheared Aanti-Aanti Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of
5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MUL
 
 | | Crystal structure of the E. coli HU alpha2 protein | | Descriptor: | DNA binding protein HU-alpha | | Authors: | Ramstein, J, Hervouet, N, Coste, F, Zelwer, C, Oberto, J, Castaing, B. | | Deposit date: | 2002-09-24 | | Release date: | 2003-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence of a Thermal Unfolding Dimeric Intermediate for the Escherichia coli Histone-like HU Proteins: Thermodynamics and Structure.
J.Mol.Biol., 331, 2003
|
|
1RL3
 
 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
4JP4
 
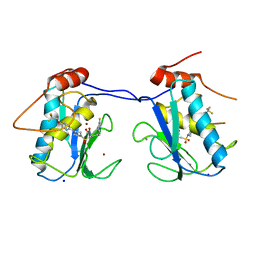 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J89
 
 | | Different photochemical events of a genetically encoded aryl azide define and modulate GFP fluorescence | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1RQ7
 
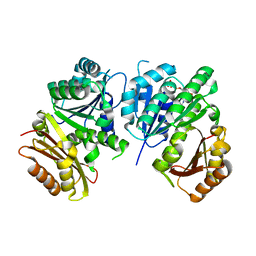 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH GDP | | Descriptor: | Cell division protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1RI1
 
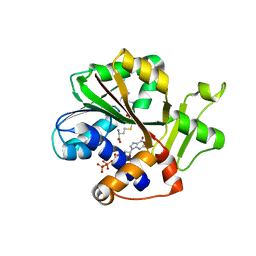 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1R2B
 
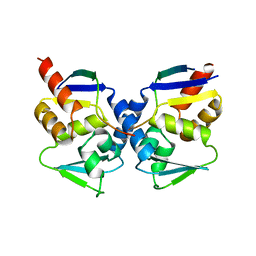 | | Crystal structure of the BCL6 BTB domain complexed with a SMRT co-repressor peptide | | Descriptor: | B-cell lymphoma 6 protein, Nuclear receptor co-repressor 2 | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
4JZ6
 
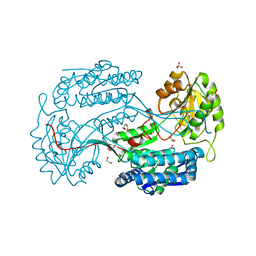 | | Crystal structure of a salicylaldehyde dehydrogenase from Pseudomonas putida G7 complexed with salicylaldehyde | | Descriptor: | 1,2-ETHANEDIOL, SALICYLALDEHYDE, SULFATE ION, ... | | Authors: | Coitinho, J.B, Nagem, R.A.P. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structural and Kinetic Properties of the Aldehyde Dehydrogenase NahF, a Broad Substrate Specificity Enzyme for Aldehyde Oxidation.
Biochemistry, 55, 2016
|
|
4E52
 
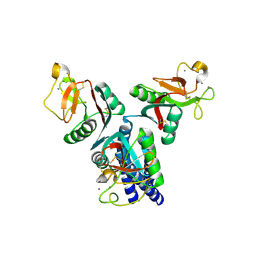 | |
1RI0
 
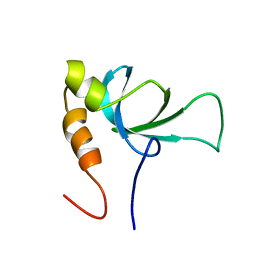 | |
1UIW
 
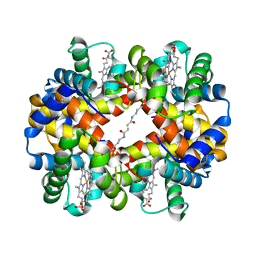 | | Crystal Structures of Unliganded and Half-Liganded Human Hemoglobin Derivatives Cross-Linked between Lys 82beta1 and Lys 82beta2 | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Park, S.-Y, Shibayama, N, Tame, J.R.H. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of unliganded and half-liganded human hemoglobin derivatives cross-linked between Lys 82beta1 and Lys 82beta2
Biochemistry, 43, 2004
|
|
4NB0
 
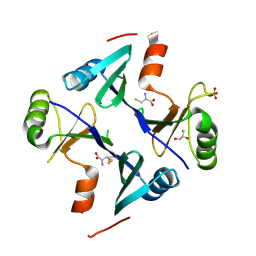 | | Crystal Structure of FosB from Staphylococcus aureus with BS-Cys9 disulfide at 1.62 Angstrom Resolution | | Descriptor: | CYSTEINE, GLYCEROL, Metallothiol transferase FosB, ... | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
1V7R
 
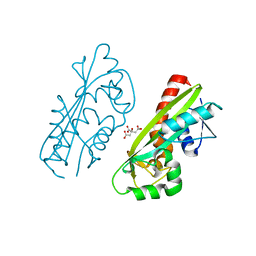 | |
4EOJ
 
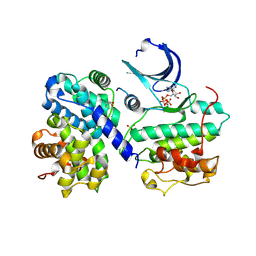 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4ES9
 
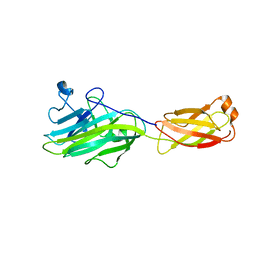 | |
