5OK2
 
 | |
8BJ3
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in complex with histidinol-phosphate | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, [(2~{S})-3-(1~{H}-imidazol-4-yl)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]propyl] dihydrogen phosphate, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
5OB9
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole (crystals grown using ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
6T4E
 
 | |
5FUL
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5OLE
 
 | |
6UCZ
 
 | |
6UFV
 
 | |
5BZW
 
 | |
5AJN
 
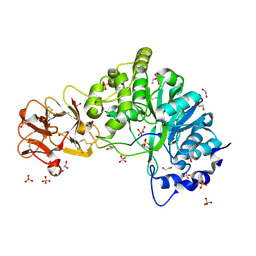 | | Crystal structure of the inactive form of GalNAc-T2 in complex with the glycopeptide MUC5AC-Cys13 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUCIN, ... | | Authors: | Lira-Navarrete, E, delasRivas, M, Companon, I, Pallares, M.C, Kong, Y, Iglesias-Fernandez, J, Bernardes, G.J.L, Peregrina, J.M, Rovira, C, Bernado, P, Bruscolini, P, Clausen, H, Lostao, A, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Nat.Commun., 6, 2015
|
|
6JOM
 
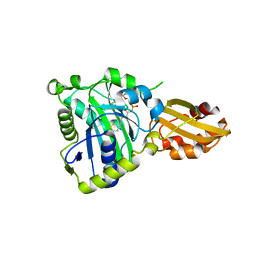 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
8ZKT
 
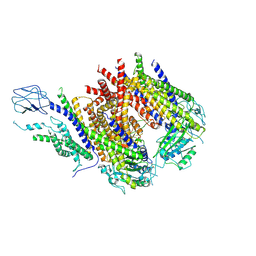 | |
8ZKE
 
 | | Cryo-EM structure of inward-facing Anhydromuropeptide permease (AmpG) in complex with GlcNAc-1,6-anhMurNAc | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, Muropeptide transporter | | Authors: | Chang, N, Kim, U, Yoo, Y, Kim, H, Cho, H. | | Deposit date: | 2024-05-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural and functional insights of AmpG in muropeptide transport and multiple beta-lactam antibiotics resistance.
Nat Commun, 16, 2025
|
|
8ZKU
 
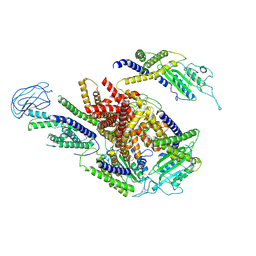 | |
8ZNU
 
 | | Catalytic antibody T99_C220A_dP95 | | Descriptor: | 1,2-ETHANEDIOL, CACODYLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Tsuyuguchi, M, Kato, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical differences between non-catalytic and catalytic antibodies.
Mabs, 17, 2025
|
|
8ZNT
 
 | | Catalytic antibody T99_C220A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Catalytic antibody T99_C220A, ... | | Authors: | Kobayashi, J, Yoshida, H, Tsuyuguchi, M, Kato, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and biochemical differences between non-catalytic and catalytic antibodies.
Mabs, 17, 2025
|
|
6J5L
 
 | |
5LOM
 
 | | Crystal structure of the PBP SocA from Agrobacterium tumefaciens C58 in complex with DFG at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Deoxyfructosyl-amino Acid Transporter Periplasmic Binding Protein, Deoxyfructosylglutamine | | Authors: | Marty, L, Vigouroux, A, Morera, S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
1P3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
5I80
 
 | | BRD4 in complex with Cpd2 (N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4501 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
7Y3Y
 
 | | Crystal structure of BTG13 mutant (T299V) | | Descriptor: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | Authors: | Hou, X.D, Fu, K, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3X
 
 | | Crystal structure of BTG13 mutant (H58F) | | Descriptor: | FE (III) ION, GLYCEROL, Questin oxidase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3W
 
 | |
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
4IV5
 
 | |
