6P69
 
 | |
6P6K
 
 | |
6YYO
 
 | | Structure of Cathepsin S in complex with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylsulfonylpiperazin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
8I2L
 
 | |
6P8Z
 
 | | Crystal structure of human KRAS G12C covalently bound to an acryloylazetidine acetamide inhibitor | | Descriptor: | 2-[5-chloro-2-cyclopropyl-3-(5-methoxy-3,4-dihydroisoquinoline-2(1H)-carbonyl)-7-methyl-1H-indol-1-yl]-N-(1-propanoylazetidin-3-yl)acetamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery ofN-(1-Acryloylazetidin-3-yl)-2-(1H-indol-1-yl)acetamides as Covalent Inhibitors of KRASG12C.
Acs Med.Chem.Lett., 10, 2019
|
|
6Z0R
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with fragment 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanylpyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.308 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
8ERI
 
 | |
8RJ7
 
 | |
8I5S
 
 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
8RBY
 
 | |
8I5R
 
 | |
6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
8D6D
 
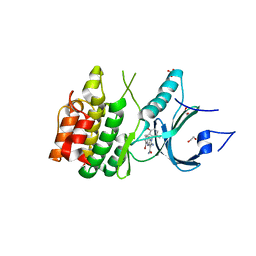 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
6WE9
 
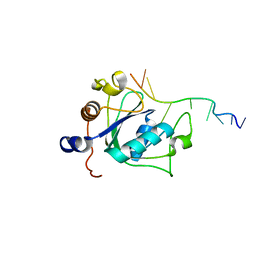 | | YTH domain of human YTHDC1 with 11mer ssDNA Containing N6mA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*GP*(6MA)P*CP*TP*CP*TP*G)-3'), GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
8F1Y
 
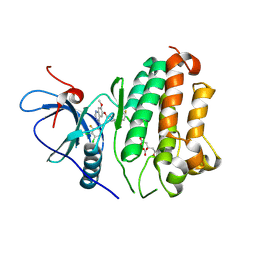 | | EGFR kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1W
 
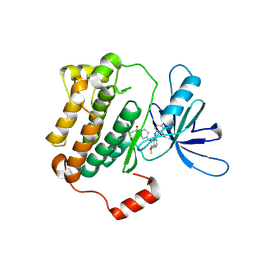 | | EGFR(T790M/V948R) kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6ZLP
 
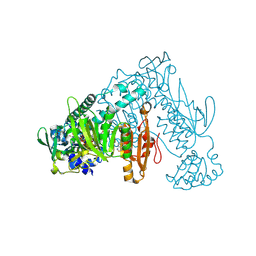 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 4-Aminopiazthiole | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
4XJR
 
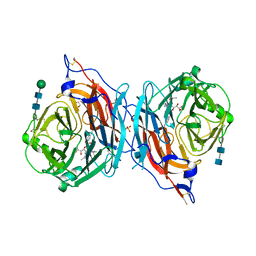 | | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed | | Descriptor: | (6R)-2,6-anhydro-3,4,5-trideoxy-6-[(2S)-2,3-dihydroxypropanoyl]-3-fluoro-5-[(2-methylpropanoyl)amino]-4-triaza-1,2-dien -2-ium-1-yl-L-gulonic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I, Guillon, P, Carroux, C, Chavas, L, von Itzstein, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6WQV
 
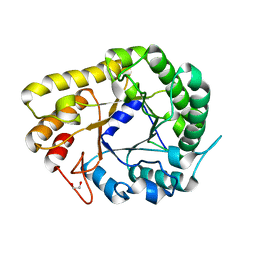 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
8DG6
 
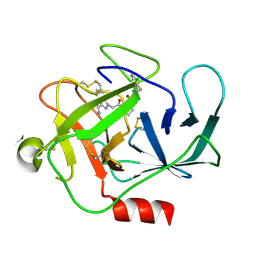 | |
6IGH
 
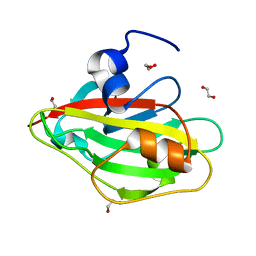 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6XHN
 
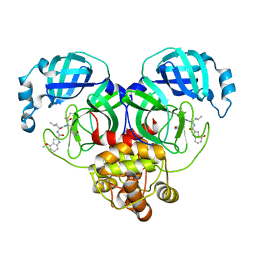 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
8KGA
 
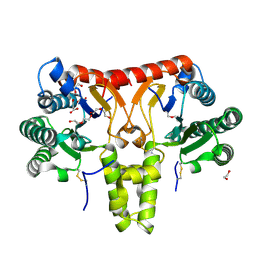 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
8RI4
 
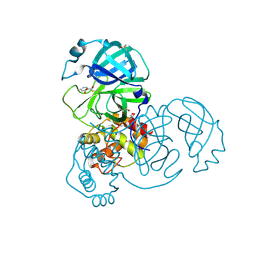 | | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone | | Descriptor: | 3C-like proteinase nsp5, 4-METHYLBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Charton, J, Deprez, B, Hanoulle, X. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone
To Be Published
|
|
