8EPR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5TCY
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
8OQR
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-80 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-cyanobenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OXN
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8EUK
 
 | |
7M5I
 
 | | Endolysin from Escherichia coli O157:H7 phage FAHEc1 | | Descriptor: | Endolysin, PHOSPHATE ION | | Authors: | Love, M.J, Coombes, D, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Viruses, 13, 2021
|
|
5NEM
 
 | |
3NWT
 
 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
1S78
 
 | | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, ... | | Authors: | Franklin, M.C, Carey, K.D, Vajdos, F.F, Leahy, D.J, de Vos, A.M, Sliwkowski, M.X. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Cancer Cell, 5, 2004
|
|
5TDT
 
 | |
6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
5TEN
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Vibrio vulnificus with 2,5 Furan Dicarboxylic and NADH with Intact Polyhistidine Tag | | Descriptor: | 2,5 Furan Dicarboxylic Acid, 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mank, N, Pote, S, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
3CD1
 
 | |
5NFR
 
 | | Crystal structure of malate dehydrogenase from Plasmodium falciparum (PfMDH) | | Descriptor: | CITRIC ACID, Malate dehydrogenase | | Authors: | Lunev, S, Romero, A.R, Batista, F.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-03-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oligomeric interfaces as a tool in drug discovery: Specific interference with activity of malate dehydrogenase of Plasmodium falciparum in vitro.
PLoS ONE, 13, 2018
|
|
3NXT
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E-and Z-isomers of a Series of 5-substituted 2,4-diaminofuro[2m,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(E)-2-cyclopropyl-2-(2-methoxyphenyl)ethenyl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1I09
 
 | |
5TEK
 
 | | Apo Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Mycobacterium tuberculosis | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Mank, N, Pote, S, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
8EXD
 
 | |
6HD7
 
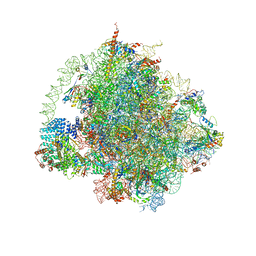 | | Cryo-EM structure of the ribosome-NatA complex | | Descriptor: | 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Becker, T, Beckmann, R. | | Deposit date: | 2018-08-17 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
8F11
 
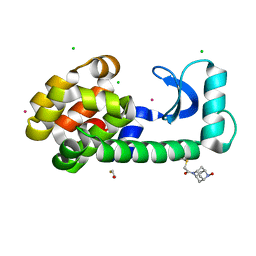 | | T4 lysozyme with a 2,6-diazaadamantane nitroxide (DZD) spin label | | Descriptor: | 1-[(1r,3r,5r,7r)-6-hydroxy-2,6-diazatricyclo[3.3.1.1~3,7~]decan-2-yl]ethan-1-one, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wilson, M.A, Madzelan, P, Rajca, A, Stein, R, Yang, Z. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Cucurbit[7]uril Enhances Distance Measurements of Spin-Labeled Proteins.
J.Am.Chem.Soc., 145, 2023
|
|
5NGT
 
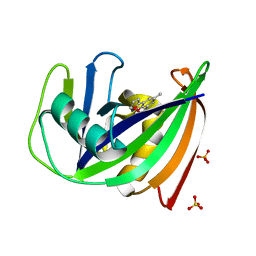 | | Crystal structure of human MTH1 in complex with inhibitor 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
8EPO
 
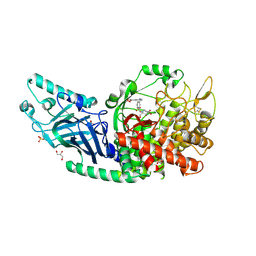 | | Co-crystal structure of Chaetomium glucosidase with compound 18 | | Descriptor: | (3P)-3-(5,6-dihydro-1,4-dioxin-2-yl)-5-{[(3-{[(2R,3R,4R,5S)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidin-1-yl]methyl}phenyl)methyl]amino}benzonitrile, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EL3
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a loose interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
1KO9
 
 | | Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG1 | | Descriptor: | 8-oxoguanine DNA glycosylase, SULFATE ION | | Authors: | Bjoras, M, Seeberg, E, Luna, L, Pearl, L.H, Barrett, T.E. | | Deposit date: | 2001-12-20 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase.
J.Mol.Biol., 317, 2002
|
|
5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
