7KZG
 
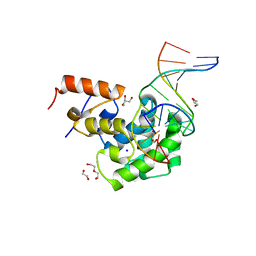 | | Human MBD4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
7KZ1
 
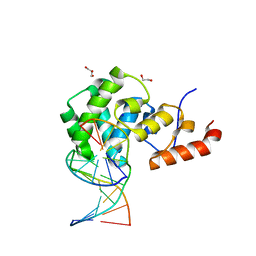 | | Human MBD4 glycosylase domain bound to DNA containing an abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Pidugu, L.S, Bright, H, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
6GIT
 
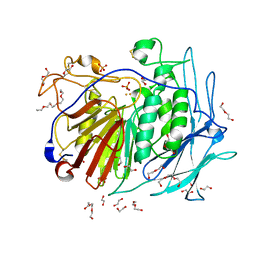 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GNL
 
 | |
6GO9
 
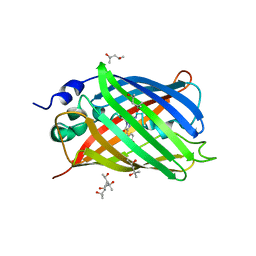 | | Structure of GFPmut2 crystallized at pH 6 and transferred to pH 7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Lolli, G, Raboni, S, Pasqualetto, E, Campanini, B, Mozzarelli, A, Bettati, S, Battistutta, R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | Insight into GFPmut2 pH Dependence by Single Crystal Microspectrophotometry and X-ray Crystallography.
J.Phys.Chem.B, 122, 2018
|
|
7KX4
 
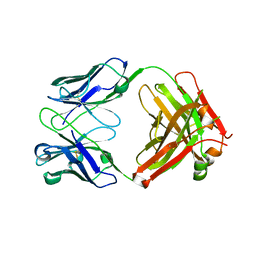 | | Anti-CCHFV ADI-36121 Fab | | Descriptor: | ADI-36121 Fab heavy chain, ADI-36121 Fab light chain | | Authors: | Mishra, A.K, McLellan, J.S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of synergistic neutralization of Crimean-Congo hemorrhagic fever virus by human antibodies.
Science, 375, 2022
|
|
6GFV
 
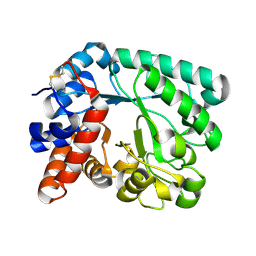 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
7T15
 
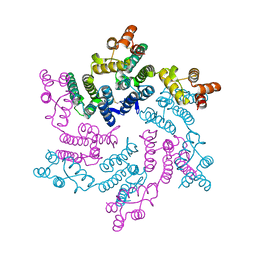 | | Hexameric SIVcpz CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, Dickson, C.F, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
7T14
 
 | | Hexameric SIVmac CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
7T31
 
 | | X-ray Structure of Clostridiodies difficile PilW | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Putative pilin protein chimera | | Authors: | Ronish, L.A, Piepenbrink, K.H. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
J.Biol.Chem., 298, 2022
|
|
7T99
 
 | |
7KQK
 
 | |
6GP5
 
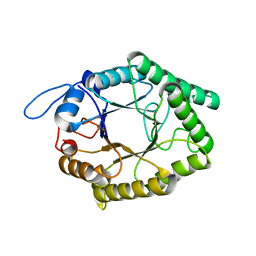 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
6GZE
 
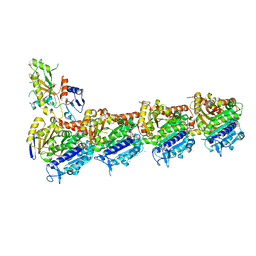 | | Tubulin-GDP.BeF complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2018-07-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural model for differential cap maturation at growing microtubule ends.
Elife, 9, 2020
|
|
7LO4
 
 | |
6HC1
 
 | |
6HFA
 
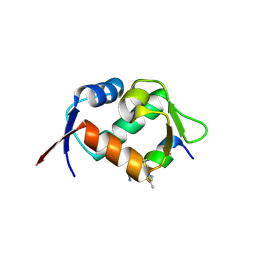 | | Crystal structure of hDM2 in complex with a C-terminal triurea capped peptide chimera foldamer. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, LM266, 1-[(2~{S})-2-azanyl-3-methyl-butyl]urea | | Authors: | Buratto, J, Mauran, L, Goudreau, S, Fribourg, S, Guichard, G. | | Deposit date: | 2018-08-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 2020
|
|
6HGU
 
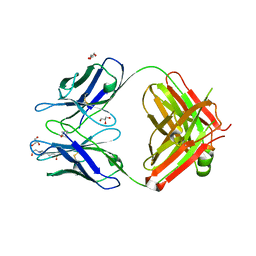 | | Crystal Structure of an anti-APP-tag Fab | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, anti-APP-tag Fab heavy-chain, ... | | Authors: | Rondeau, J.M, Goepfert, A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis Reveals that the Cytokine IL-17F Forms a Homodimeric Complex with Receptor IL-17RC to Drive IL-17RA-Independent Signaling.
Immunity, 52, 2020
|
|
7LGD
 
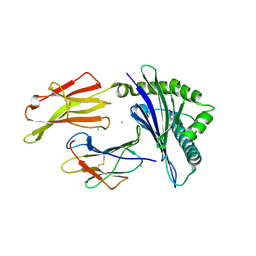 | | HLA-B*07:02 in complex with SARS-CoV-2 nucleocapsid peptide N105-113 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7LUN
 
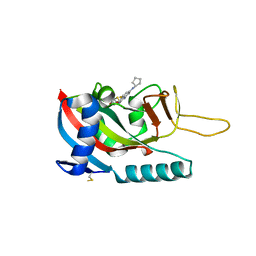 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN011980 | | Descriptor: | 7-(cyclopentylamino)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7LGT
 
 | | HLA-B*07:02 in complex with 229E-derived coronavirus nucleocapsid peptide N75-83 | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
6HOT
 
 | |
7LOG
 
 | | T4 lysozyme mutant L99A in complex with 3-butylpyridine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-butylpyridine, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6I0R
 
 | | Structure of quinolinate synthase in complex with 5-mercaptopyridine-2,3-dicarboxylic acid | | Descriptor: | 5-mercaptopyridine-2,3-dicarboxylic acid, FE (III) ION, HYDROSULFURIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
7LXA
 
 | | T4 lysozyme mutant L99A | | Descriptor: | (2-methylprop-2-en-1-yl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
