4X51
 
 | |
4WW5
 
 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW9
 
 | |
4WWM
 
 | | X-ray crystal structure of Sulfolobus solfataricus Urm1 | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Bray, S.M, Anjum, S.R, Blackwood, J.K, Kilkenny, M.L, Coelho, M.A, Foster, B.M, Pellegrini, L, Robinson, N.P. | | Deposit date: | 2014-11-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius.
Nat Commun, 6, 2015
|
|
4X6H
 
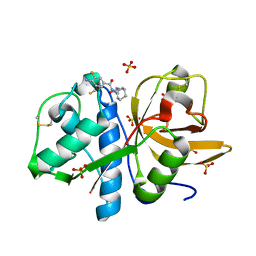 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | Descriptor: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | Authors: | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
4WXY
 
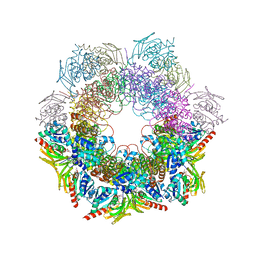 | |
4X8D
 
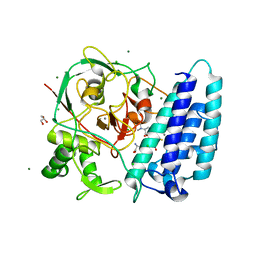 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB in complex with N,N-dimethyl-histidine and gamma-glutamyl-cysteine | | Descriptor: | CALCIUM ION, CHLORIDE ION, GAMMA-GLUTAMYLCYSTEINE, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7ZKP
 
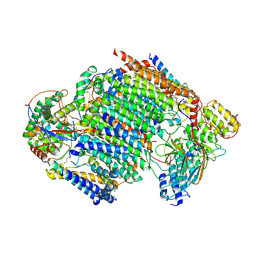 | | Late assembly intermediate of the proximal proton pumping module of complex I with assembly factors NDUFAF1 and CIA84 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, ... | | Authors: | Schiller, J, Laube, E, Vonck, J, Zickermann, V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-23 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling.
Sci Adv, 8, 2022
|
|
7ZG7
 
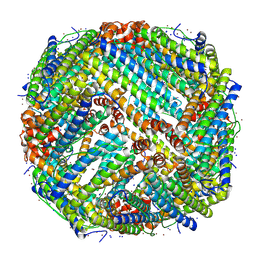 | | Structure of human Apoferritin obtained from ssDNA coated grid | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Hrebik, D, Plevka, P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | Polyelectrolyte coating of cryo-EM grids improves lateral distribution and prevents aggregation of macromolecules.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZXW
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 5-(morpholin-4-ylmethyl)quinolin-8-ol, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Cornu, J.D, Ibba, R, Roversi, P, Zitzmann, N. | | Deposit date: | 2022-05-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 9, 2022
|
|
5OSC
 
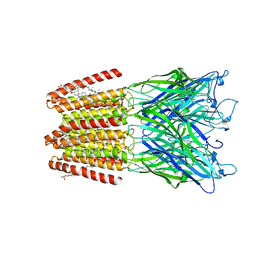 | | GLIC-GABAAR alpha1 chimera crystallized in complex with pregnenolone sulfate at pH 4.5 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Laverty, D.C, Gold, M.G, Smart, T.G. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7ZR0
 
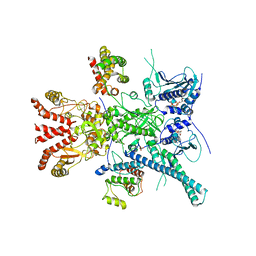 | | CryoEM structure of HSP90-CDC37-BRAF(V600E) complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Oberoi, J, Pearl, L.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation.
Nat Commun, 13, 2022
|
|
7ZR5
 
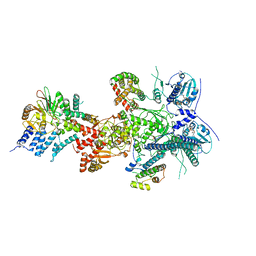 | |
5OYA
 
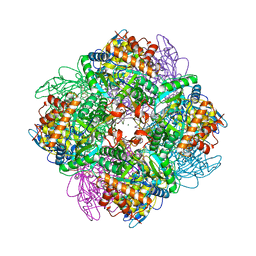 | | Unusual posttranslational modifications revealed in crystal structures of diatom Rubisco. | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Valegard, K, Andersson, I. | | Deposit date: | 2017-09-08 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications.
J. Biol. Chem., 293, 2018
|
|
7ZLD
 
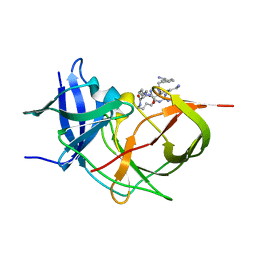 | | Crystal Structure of Unlinked NS2B_NS3 Protease from Zika Virus in Complex with Inhibitor MI-2223 | | Descriptor: | (2~{S})-2-[2-[3-(aminomethyl)phenyl]ethanoylamino]-6-azanyl-~{N}-[(2~{S})-6-azanyl-1-[[(5~{R})-6-azanyl-5-carbamimidamido-6-oxidanylidene-hexyl]amino]-1-oxidanylidene-hexan-2-yl]hexanamide, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Thermodynamic characterization of a macrocyclic Zika virus NS2B/NS3 protease inhibitor and its acyclic analogs.
Arch Pharm, 356, 2023
|
|
7ZKG
 
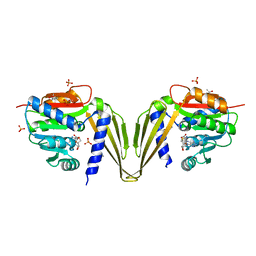 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
7ZPM
 
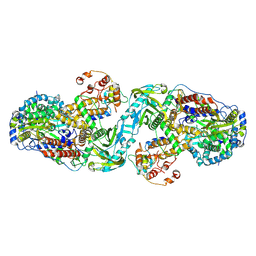 | | Influenza A/H7N9 polymerase apo-protein dimer complex | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106.
Cell Rep, 42, 2023
|
|
7ZR6
 
 | |
7ZPL
 
 | |
7ZLC
 
 | |
7ZMI
 
 | |
7ZZQ
 
 | |
5OYH
 
 | | crystal structure of the catalytic core of a rhodopsin-guanylyl cyclase with converted specificity in complex with ATPalphaS | | Descriptor: | ADENOSINE-5'-SP-ALPHA-THIO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Broser, M, Scheib, U, Hegemann, P. | | Deposit date: | 2017-09-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 angstrom structure of the adenylyl cyclase domain.
Nat Commun, 9, 2018
|
|
8A00
 
 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
7ZNY
 
 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
