8BSW
 
 | |
8RUR
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 2 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [1.38 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8BSV
 
 | |
1VZU
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
6ZGV
 
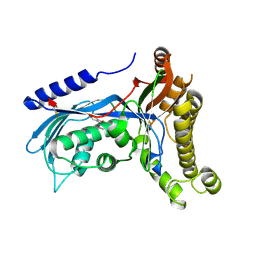 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 2-(4-chlorophenyl)-~{N}-pyrimidin-2-yl-ethanamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
7RC9
 
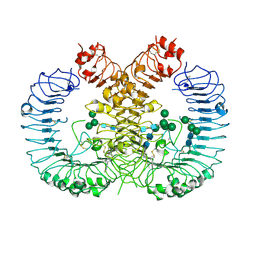 | | Crystal structure of human TLR8 ectodomain bound to small molecule antagonist 21 | | Descriptor: | 2-(2,6-dimethylpyridin-4-yl)-5-(piperidin-4-yl)-3-(propan-2-yl)-1H-indole, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Critton, D.A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Identification of 2-Pyridinylindole-Based Dual Antagonists of Toll-like Receptors 7 and 8 (TLR7/8).
Acs Med.Chem.Lett., 13, 2022
|
|
8W6P
 
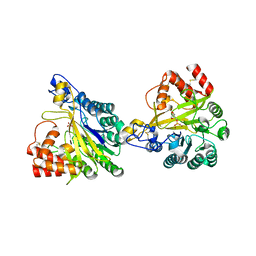 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
6OMP
 
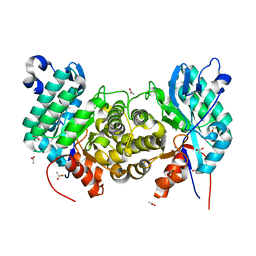 | | Crystal structure of apo PtmU3 | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PtmU3 | | Authors: | Liu, Y.C, Dong, L.B, Shen, B. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and Crystal Structure of a Nonheme Diiron Monooxygenase Involved in Platensimycin and Platencin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
8W6R
 
 | | murine SMPDL3A bound to sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
8BKH
 
 | |
8BKN
 
 | |
6OP1
 
 | |
6O82
 
 | | S. pombe ubiquitin E1 complex with a ubiquitin-AMP mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-(sulfamoylamino)adenosine, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
8RK0
 
 | | HCV E1/E2 homodimer complex, ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HCV E1, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 2024
|
|
6OSX
 
 | | Crystal structure of uncharacterized protein ECL_02694 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Protein YmbA, ... | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of uncharacterized protein ECL_02694
To Be Published
|
|
6O9O
 
 | | Crystal Structure of SMYD3 with Potent and Selective Isoxazole Amide Inhibitor 1 | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors
To be published
|
|
8J5J
 
 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
6O9T
 
 | | KirBac3.1 mutant at a resolution of 4.1 Angstroms | | Descriptor: | 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
8RJJ
 
 | | HCV E1/E2 homodimer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 2024
|
|
6O9V
 
 | | KirBac3.1 mutant at a resolution of 3.1 Angstroms | | Descriptor: | 1,1-Methanediyl Bismethanethiosulfonate, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
8J6R
 
 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
5IGX
 
 | |
8J6Q
 
 | | Cryo-EM structure of the 3-HB and compound 9n-bound human HCAR2-Gi1 complex | | Descriptor: | (3R)-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8IUN
 
 | | Cryo-EM structure of the CRT-LESS RC-LH core complex from roseiflexus castenholzii | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-03-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
