5N7M
 
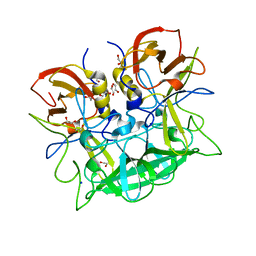 | |
1NV4
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate, EDTA and Thallium (1 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-02 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
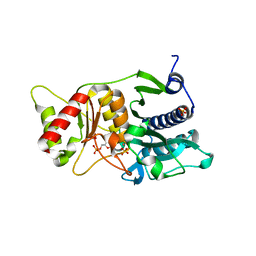
7R84
 
 | |
6O55
 
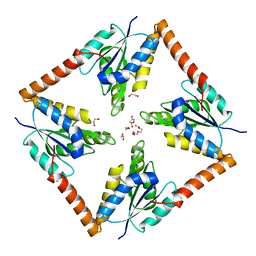 | |
6O49
 
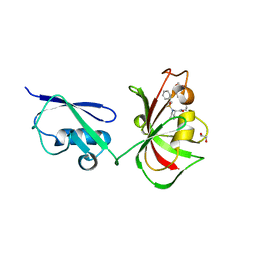 | |
6O5M
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 10bb | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
J.Med.Chem., 62, 2019
|
|
8RP8
 
 | | Structure of K2 Fab in complex with human CD47 ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Laursen, M, Kelpsas, V, Rose, N. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
Mabs, 16, 2024
|
|
3JPW
 
 | | Crystal structure of amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
6OCQ
 
 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | Descriptor: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
8SBU
 
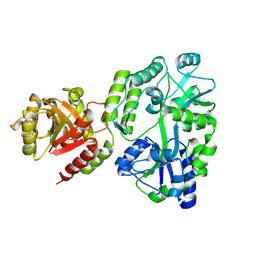 | | Crystal structure of MBP fusion with HPPK from Methanocaldococcus jannaschii | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Shaw, G.X, Needle, D, Stair, N.R, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-04-04 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of MBP fusion with HPPK from Methanocaldococcus jannaschii
To be published
|
|
6OHK
 
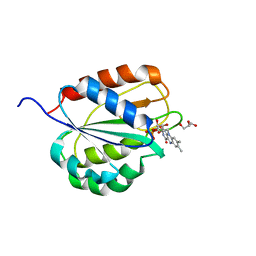 | |
6ZGO
 
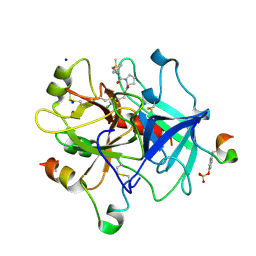 | | Thrombin in complex with D-Phe-Pro-2-chlorofuran derivative (13l) | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(5-chloranylfuran-2-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-2-chlorofuran derivative (13l)
To be published
|
|
8URK
 
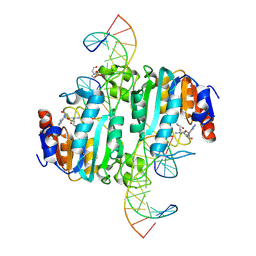 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrates | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA1, DNA2, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2023-10-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
6OM1
 
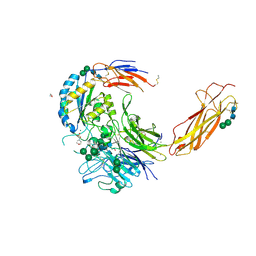 | | Crystal structure of an atypical integrin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.C, Springer, T.A. | | Deposit date: | 2019-04-17 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | General structural features that regulate integrin affinity revealed by atypical alpha V beta 8.
Nat Commun, 10, 2019
|
|
6ZFH
 
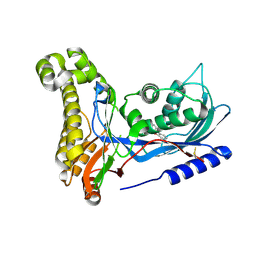 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.439 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
8RG8
 
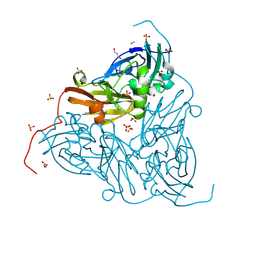 | | High pH (8.0) nitrite-bound MSOX movie series dataset 1 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [0.68 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RFY
 
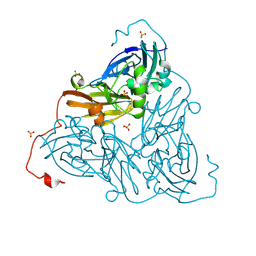 | | High pH (8.0) as-isolated MSOX movie series dataset 30 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [12.3 MGy] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6OK4
 
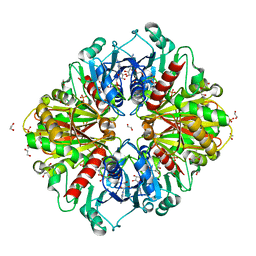 | |
8RGB
 
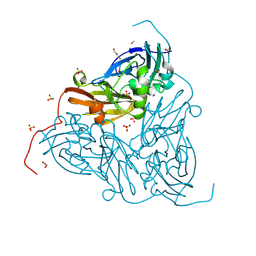 | | High pH (8.0) nitrite-bound MSOX movie series dataset 5 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [3.4 MGy] | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6OKH
 
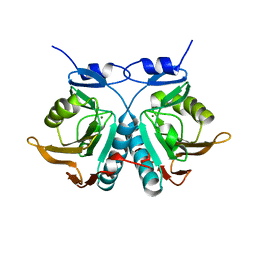 | |
6OOH
 
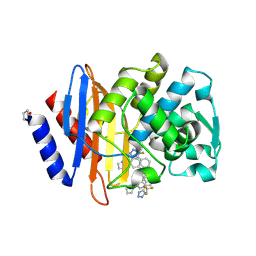 | | CTX-M-27 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
8RYJ
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 3 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [2.07 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RFU
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 40 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [24.4 MGy] - water ligand + decarboxylated AspCAT (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RFT
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 17 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [10.37 MGy] - water ligand + decarboxylated AspCAT | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RFS
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 11 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [6.71 MGy] - water ligand | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
