5GQW
 
 | |
8F69
 
 | | Crystal structure of murine PolG2 dimer bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wojtaszek, J.L, Hoff, K.E, Williams, R.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-specific roles for PolG2-DNA complexes in maintenance and replication of mitochondrial DNA.
Nucleic Acids Res., 51, 2023
|
|
2OK8
 
 | | Ferredoxin-NADP+ reductase from Plasmodium falciparum | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Putative ferredoxin--NADP reductase | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2007-01-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ferredoxin-NADP(+) Reductase from Plasmodium falciparum Undergoes NADP(+)-dependent Dimerization and Inactivation: Functional and Crystallographic Analysis.
J.Mol.Biol., 367, 2007
|
|
5D0S
 
 | |
4RN2
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,17,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
3K1F
 
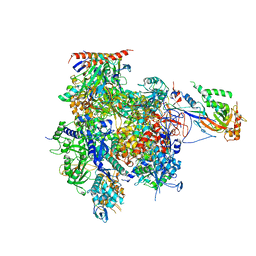 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | Deposit date: | 2009-09-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
2BGD
 
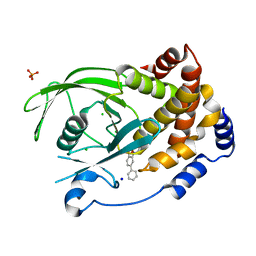 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4RQI
 
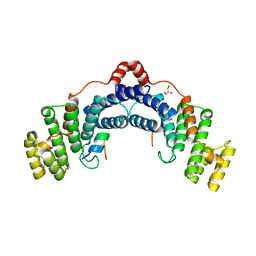 | | Structure of TRF2/RAP1 secondary interaction binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Telomeric repeat-binding factor 2, ... | | Authors: | Miron, S, Guimaraes, B, Gaullier, G, Giraud-Panis, M.-J, Gilson, E, Le Du, M.-H. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4405 Å) | | Cite: | A higher-order entity formed by the flexible assembly of RAP1 with TRF2.
Nucleic Acids Res., 44, 2016
|
|
1QJE
 
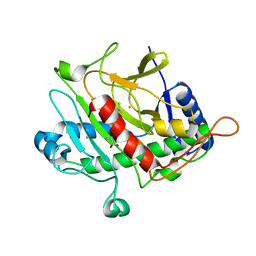 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
3K4A
 
 | |
2NXF
 
 | | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393 | | Descriptor: | ETHANOL, PHOSPHATE ION, Putative dimetal phosphatase, ... | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393
To be Published
|
|
1KRA
 
 | |
6F1U
 
 | | N terminal region of dynein tail domains in complex with dynactin filament and BICDR-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
7P4T
 
 | | Tetrameric structure of murine SapA | | Descriptor: | Saposin-A | | Authors: | Shamin, M, Deane, J.E. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A Tetrameric Assembly of Saposin A: Increasing Structural Diversity in Lipid Transfer Proteins.
Contact, 4, 2021
|
|
1GXC
 
 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
1KRB
 
 | |
2C8F
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (NAD-bound state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
1T7T
 
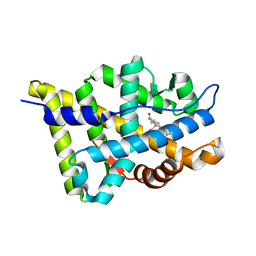 | | Crystal structure of the androgen receptor ligand binding domain in complex with 5-alpha dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1HAK
 
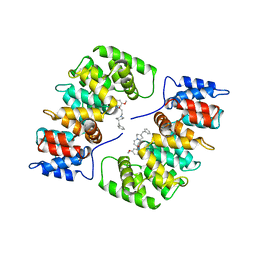 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
4RW4
 
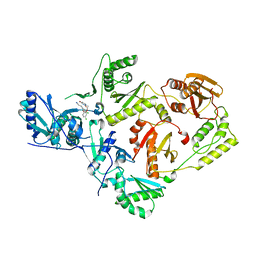 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N,Y181C) variant in complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | Descriptor: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-12-01 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.674 Å) | | Cite: | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
5DID
 
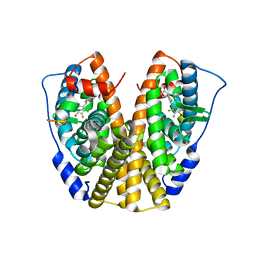 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a difluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6F9H
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | Descriptor: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6F1K
 
 | | Structure of ARTD2/PARP2 WGR domain bound to double strand DNA without 5'phosphate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*CP*TP*AP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*TP*AP*GP*GP*C)-3'), GLYCEROL, ... | | Authors: | Obaji, E, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for DNA break recognition by ARTD2/PARP2.
Nucleic Acids Res., 46, 2018
|
|
2O57
 
 | | Crystal Structure of a putative sarcosine dimethylglycine methyltransferase from Galdieria sulphuraria | | Descriptor: | putative sarcosine dimethylglycine methyltransferase | | Authors: | Mccoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Crystal Structure of a putative sarcosine dimethylglycine methyltransferase from Galdieria sulphuraria
To be Published
|
|
5D5R
 
 | | Horse-heart myoglobin - deoxy state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Barends, T, Schlichting, I. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
