6EQV
 
 | |
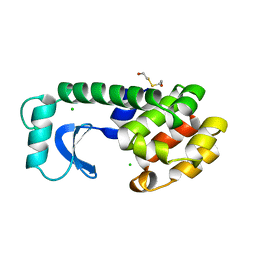
1L85
 
 | |
1T66
 
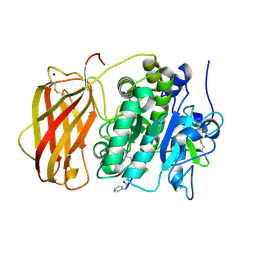
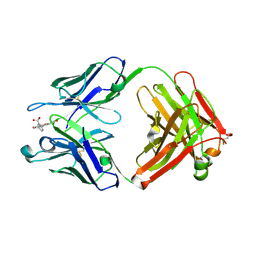 | | The structure of FAB with intermediate affinity for fluorescein. | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, immunoglobulin heavy chain, immunoglobulin light chain | | Authors: | Terzyan, S, Ramsland, P.A, Voss Jr, E.W, Herron, J.N, Edmundson, A.B. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional Structures of Idiotypically Related Fabs with Intermediate and High Affinity for Fluorescein.
J.Mol.Biol., 339, 2004
|
|
1T8Q
 
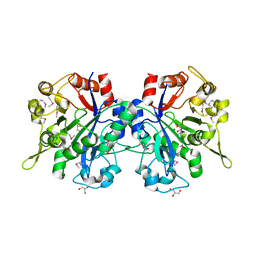 | | Structural genomics, Crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, periplasmic, ... | | Authors: | Zhang, R, Kim, Y, Dementieva, I, Duke, N, Stols, L, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli
To be Published
|
|
4JF8
 
 | |
3ZQZ
 
 | | CRYSTAL STRUCTURE OF ANCE IN COMPLEX WITH A SELENIUM ANALOGUE OF CAPTOPRIL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, SELENO-CAPTOPRIL, ... | | Authors: | Akif, M, Masuyer, G, Schwager, S.L.U, Bhuyan, B.J, Mugesh, G, Isaac, R.E, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Angiotensin I- Converting Enzyme in Complex with a Selenium Analogue of Captopril.
FEBS J., 278, 2011
|
|
6EM4
 
 | | State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | Descriptor: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
6EB4
 
 | | OhrB (Organic Hydroperoxide Resistance protein) from Chromobacterium violaceum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-04 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
8DST
 
 | |
6EBP
 
 | |
5CJ4
 
 | | Crystal Structure of Amino Acids 1562-1622 of MYH7 | | Descriptor: | Xrcc4-MYH7-(1562-1622) chimera protein | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinze, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
200L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
8EC3
 
 | |
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its apo form
To Be Published
|
|
8E5G
 
 | | Cryo-EM of A. veneficus cytochrome filament | | Descriptor: | HEME C, c-type cytochrome | | Authors: | Wang, F, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extracellular cytochrome nanowires appear to be ubiquitous in prokaryotes.
Cell, 186, 2023
|
|
5GRK
 
 | | Crystal structure of Uracil DNA glycosylase -Xanthine complex from Bradyrhizobium diazoefficiens | | Descriptor: | Blr0248 protein, XANTHINE | | Authors: | Patil, V.V, Ullas, V.C, Ahn, W, Varshney, U, Woo, E. | | Deposit date: | 2016-08-11 | | Release date: | 2017-05-03 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Uracil DNA glycosylase (UDG) activities in Bradyrhizobium diazoefficiens: characterization of a new class of UDG with broad substrate specificity
Nucleic Acids Res., 45, 2017
|
|
1CVW
 
 | | Crystal structure of active site-inhibited human coagulation factor VIIA (DES-GLA) | | Descriptor: | CALCIUM ION, COAGULATION FACTOR VIIA (HEAVY CHAIN) (DES-GLA), COAGULATION FACTOR VIIA (LIGHT CHAIN) (DES-GLA), ... | | Authors: | Kemball-Cook, G, Johnson, D.J.D, Tuddenham, E.G.D, Harlos, K. | | Deposit date: | 1999-08-24 | | Release date: | 1999-08-31 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of active site-inhibited human coagulation factor VIIa (des-Gla).
J.Struct.Biol., 127, 1999
|
|
7PB3
 
 | | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
3J9G
 
 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
4QUY
 
 | | yCP beta5-A49S-mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7ON8
 
 | |
1LE4
 
 | |
7OPU
 
 | |
6EJ9
 
 | |
