5EC2
 
 | | KcsA with V76ester+G77dA mutations | | Descriptor: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
5E66
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | Descriptor: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5SV6
 
 | |
5DL0
 
 | | Crystal structure of glucosidase II alpha subunit (Glc1Man2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
4N0U
 
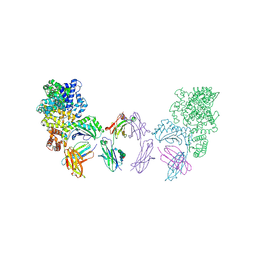 | | Ternary complex between Neonatal Fc receptor, serum albumin and Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Ig gamma-1 chain C region, ... | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2013-10-02 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into Neonatal Fc Receptor-based Recycling Mechanisms.
J.Biol.Chem., 289, 2014
|
|
4N1L
 
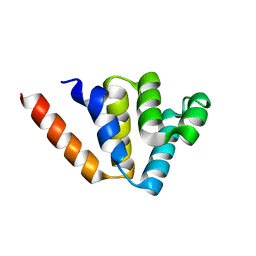 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ZGQ
 
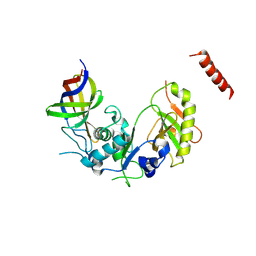 | | Structure of Cdc123 bound to eIF2-gammaDIII domain | | Descriptor: | Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
5DYP
 
 | |
4FMX
 
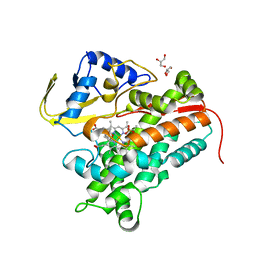 | | Crystal Structure of Substrate-Bound P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, GLYCEROL, P450cin, ... | | Authors: | Madrona, Y, Tripathi, S.M, Huiying, L, Poulos, T.L. | | Deposit date: | 2012-06-18 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
5DZF
 
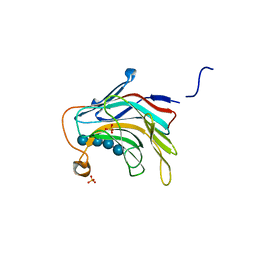 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a mixed-linkage glucan octasaccharide | | Descriptor: | SULFATE ION, beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
4D6Z
 
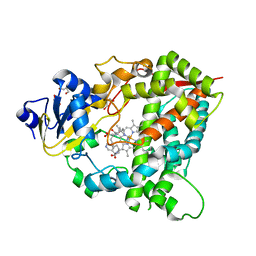 | |
4D7D
 
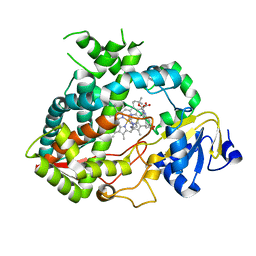 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-({3-oxo-3-[(pyridin-3-ylmethyl)amino]propyl}sulfanyl)propan-2-yl]carbamate | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
5AMY
 
 | |
4ZGN
 
 | | Structure Cdc123 complexed with the C-terminal domain of eIF2gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
4D75
 
 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl {6-oxo-6-[(pyridin-3-ylmethyl)amino]hexyl}carbamate | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
5AZF
 
 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | Authors: | Watanabe, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AWP
 
 | |
5AXG
 
 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Dextranase, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
5B2G
 
 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
5B4I
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 2) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5B2V
 
 | | Crystal Structure of P450BM3 with N-perfluorohexanoyl-L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,6-undecakis(fluoranyl)hexanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
4CQB
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
5B3K
 
 | |
5B7V
 
 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
5ANT
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 2-(2-methoxyethoxy)-6-(methylamino)-9-(phenylmethyl)-7H-purin-8-one, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
