2WF8
 
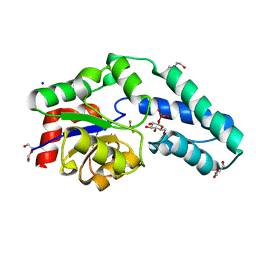 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
8SAK
 
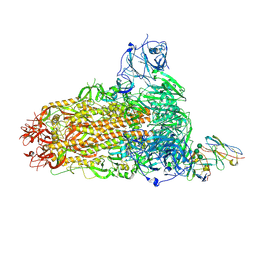 | | BtCoV-422 in complex with neutralizing antibody JC57-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McFadden, E, McLellan, J.S. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A MERS-CoV antibody neutralizes a pre-emerging group 2c bat coronavirus.
Sci Transl Med, 15, 2023
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
8S9G
 
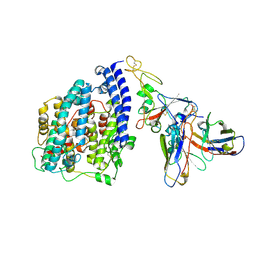 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6ZJT
 
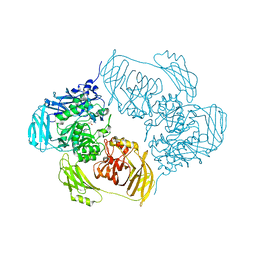 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactulose | | Descriptor: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
7QQB
 
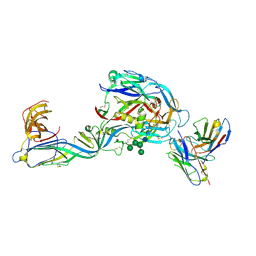 | | Crystal structure of the envelope glycoprotein complex of Puumala virus in complex with the scFv fragment of the broadly neutralizing human antibody ADI-42898 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, GLYCEROL, ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2022-01-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody.
Sci Transl Med, 15, 2023
|
|
8RBN
 
 | |
6NWX
 
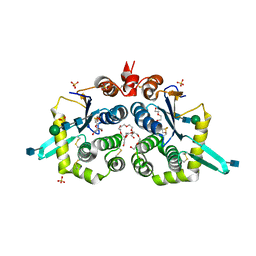 | | Structure of mouse GILT, an enzyme involved in antigen processing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-interferon-inducible lysosomal thiol reductase, PENTAETHYLENE GLYCOL, ... | | Authors: | Li, Y. | | Deposit date: | 2019-02-07 | | Release date: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gamma-interferon-inducible lysosomal thiol reductase (GILT). Maturation, activity, and mechanism of action
To Be Published
|
|
8SGM
 
 | |
8SGN
 
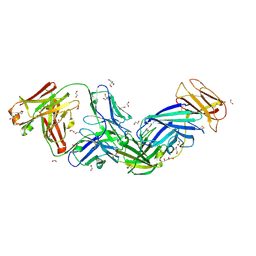 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
7OR1
 
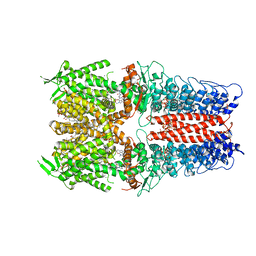 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
8SGB
 
 | |
7OR0
 
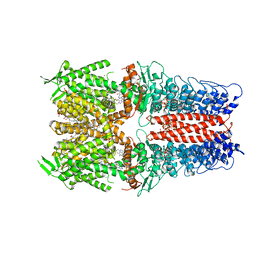 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
8SJK
 
 | | Pembrolizumab Caffeine crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY HEAVY CHAIN, ANTIBODY LIGHT CHAIN, ... | | Authors: | Larpent, P, Codan, L, Bothe, J.R, Stueber, D, Reichert, P, Fischmann, T, Su, Y, Pabit, S, Gupta, S, Iuzzolino, L, Cote, A. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small-Angle X-ray Scattering as a Powerful Tool for Phase and Crystallinity Assessment of Monoclonal Antibody Crystallites in Support of Batch Crystallization.
Mol Pharm., 21, 2024
|
|
6Z7C
 
 | | Variant Surface Glycoprotein VSGsur mutant H122A | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7E
 
 | |
6Z7D
 
 | | Variant Surface Glycoprotein VSGsur mutant H122A soaked in 0.77 mM Suramin. | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6HAQ
 
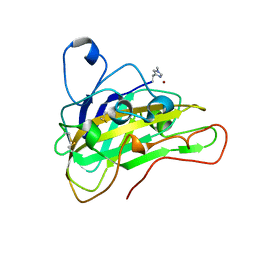 | | AFGH61B WILD-TYPE COPPER LOADED | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N, Weihe, C.D. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
8SC8
 
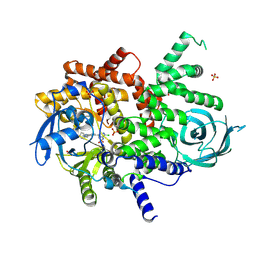 | | Structure of PI3KG in complex with MTX-531 | | Descriptor: | N-[(5P)-2-chloro-5-(4-{[(1R)-1-phenylethyl]amino}quinazolin-6-yl)pyridin-3-yl]methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whitehead, C.E, Leopold, J. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structure of PI3KG in complex with MTX-531
To Be Published
|
|
7P2Z
 
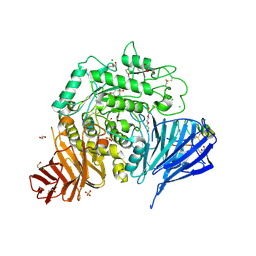 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 4 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
6O3I
 
 | | Crystal Structure of Human IDO1 bound to navoximod (NLG-919) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, trans-4-{(1R)-2-[(5S)-6-fluoro-5H-imidazo[5,1-a]isoindol-5-yl]-1-hydroxyethyl}cyclohexan-1-ol | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Clinical Candidate (1R,4r)-4-((R)-2-((S)-6-Fluoro-5H-imidazo[5,1-a]isoindol-5-yl)-1-hydroxyethyl)cyclohexan-1-ol (Navoximod), a Potent and Selective Inhibitor of Indoleamine 2,3-Dioxygenase 1.
J.Med.Chem., 62, 2019
|
|
8SAW
 
 | | CryoEM structure of DH270.UCA.G57R-CH848.10.17DT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp120A, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
6ZBW
 
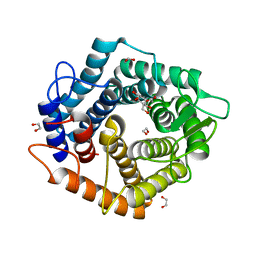 | | Structure of the D125N mutant of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, Alpha-1,6-mannanase, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
8SAN
 
 | | CryoEM structure of VRC01-CH848.0836.10 | | Descriptor: | CH848.0836.10 gp120, CH848.0836.10 gp41, VCR01 variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAU
 
 | | CryoEM structure of DH270.4-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
