6XN1
 
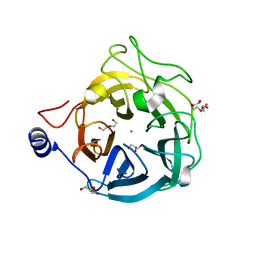 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
5Y0P
 
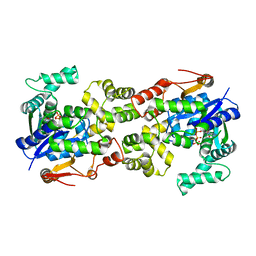 | |
5Y09
 
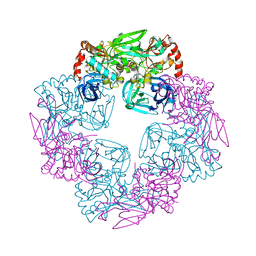 | |
8B6E
 
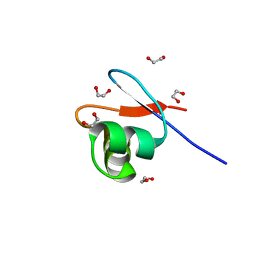 | | crystal structure of the DNA-binding short chromatophore-targeted protein sCTP-23166 from Paulinella chromatophora | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, sCTP-23166 | | Authors: | Macorano, L, Applegate, V, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | DNA-binding and protein structure of nuclear factors likely acting in genetic information processing in the Paulinella chromatophore.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6XPB
 
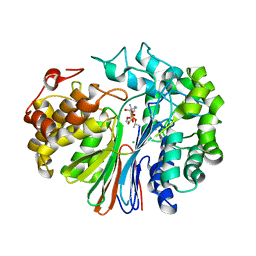 | |
5Y1U
 
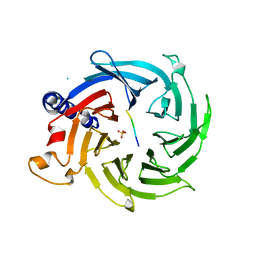 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
5Y2I
 
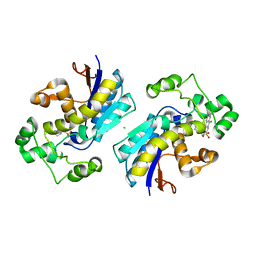 | | Phosphoglycerate mutase 1 (PGAM1) complexed with its inhibitor PGMI-004A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,4-bis(oxidanyl)-9,10-bis(oxidanylidene)-~{N}-[4-(trifluoromethyl)phenyl]anthracene-2-sulfonamide, CHLORIDE ION, ... | | Authors: | Jiang, L, Zhou, L. | | Deposit date: | 2017-07-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Phosphoglycerate mutase 1 (PGAM1) complexed with its inhibitor PGMI-004A
To Be Published
|
|
5Y2U
 
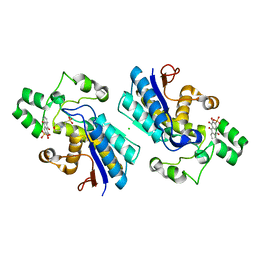 | |
5Y4E
 
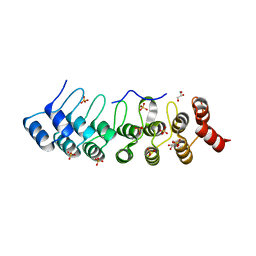 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | Descriptor: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y38
 
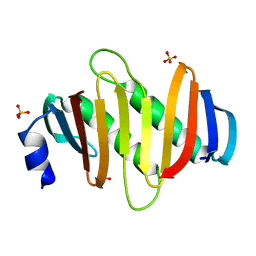 | | Crystal structure of C7orf59-HBXIP complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
5Y50
 
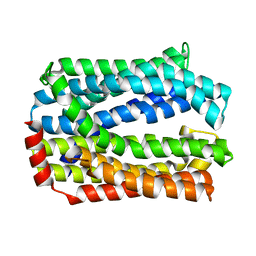 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
6O4Z
 
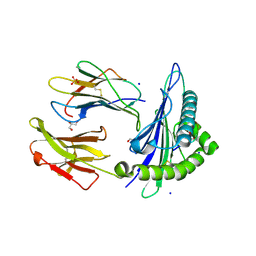 | | Structure of HLA-A2:01 with peptide MM92 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O51
 
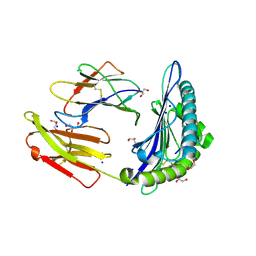 | | Structure of HLA-A2:01 with peptide MM90 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y83
 
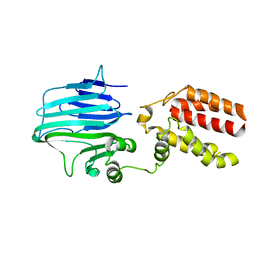 | | Crystal structure of YidC from Thermotoga maritima | | Descriptor: | Membrane protein insertase YidC | | Authors: | Huang, Y, Xin, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.842 Å) | | Cite: | Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion
FASEB J., 32, 2018
|
|
6O6O
 
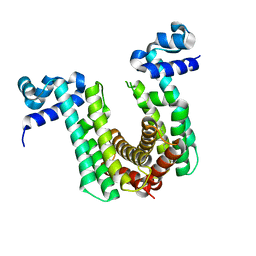 | | Structure of the regulator FasR from Mycobacterium tuberculosis | | Descriptor: | MYRISTIC ACID, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
5Y8R
 
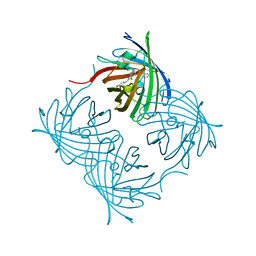 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6O69
 
 | |
5YFB
 
 | | Crystal structure of a new DPP III family member | | Descriptor: | 1,2-ETHANEDIOL, Dipeptidyl peptidase 3, GLYCEROL, ... | | Authors: | Xu, T, Sun, Z, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5Y32
 
 | | Crystal structure of PTP delta Ig1-Ig2 in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
5YG0
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS657 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | to be published
to be published
|
|
5Y59
 
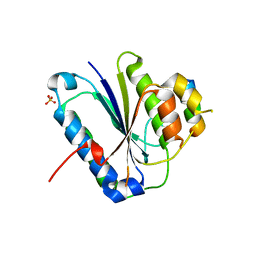 | | Crystal structure of Ku80 and Sir4 | | Descriptor: | ATP-dependent DNA helicase II subunit 2, SULFATE ION, Sir4p | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
5Y5F
 
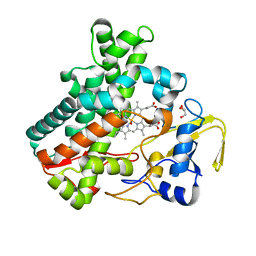 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y17
 
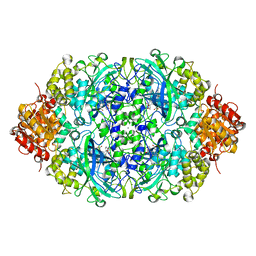 | | CATPO mutant - E316F | | Descriptor: | CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase | | Authors: | Karakus Yuzugullu, Y, Goc, G, Balci, S, Pearson, A.R, Yorke, B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5Y39
 
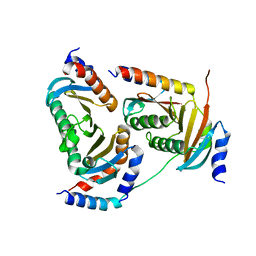 | | Crystal structure of Ragulator complex (p18 76-145) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
