7P0G
 
 | |
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
7P0C
 
 | |
4J39
 
 | |
3WF2
 
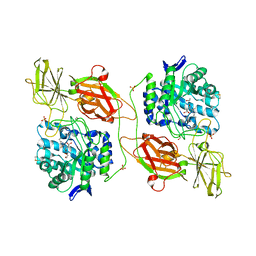 | | Crystal structure of human beta-galactosidase in complex with NBT-DGJ | | Descriptor: | (2R,3S,4R,5S)-N-butyl-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
4IF5
 
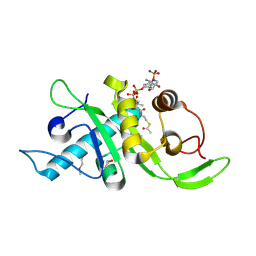 | | Structure of human Mec17 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, CHLORIDE ION | | Authors: | Davenport, A.M, Collins, L, Minor, P, Sternberg, P, Hoelz, A. | | Deposit date: | 2012-12-14 | | Release date: | 2014-05-28 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the alpha-Tubulin Acetyltransferase MEC-17.
J.Mol.Biol., 426, 2014
|
|
4ILQ
 
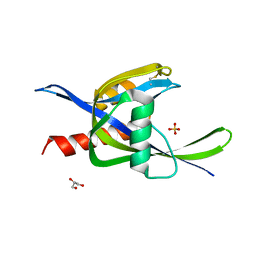 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4IJW
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
3WK2
 
 | |
7P9Q
 
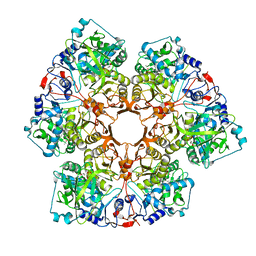 | | Crystal structure of Indole 3-Carboxylic acid decarboxylase from Arthrobacter nicotianae FI1612 in complex with co-factor prFMN. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, AnInD, MANGANESE (II) ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and biochemical characterization of the prenylated flavin mononucleotide-dependent indole-3-carboxylic acid decarboxylase.
J.Biol.Chem., 298, 2022
|
|
3WVG
 
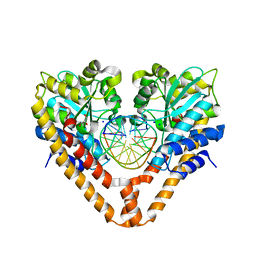 | | Time-Resolved Crystal Structure of HindIII with 0sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, SODIUM ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WPH
 
 | |
4M09
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W146Y R173Q | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Gysel, K, Hagmueller, A, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
8BDO
 
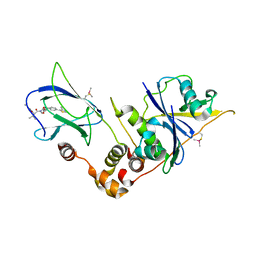 | | VCB in complex with compound 21 | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-~{N}-[4-(4-methyl-1,3-thiazol-5-yl)phenoxy]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
4IN0
 
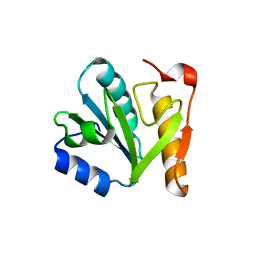 | |
4INA
 
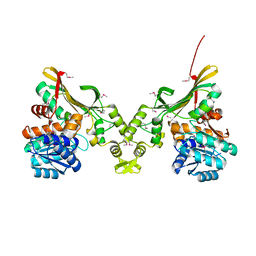 | | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes. Northeast Structural Genomics Consortium Target WsR35 | | Descriptor: | saccharopine dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes.
To be Published
|
|
8BDL
 
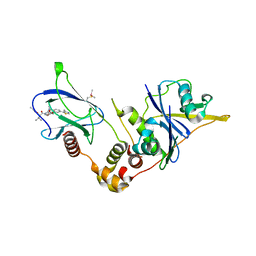 | | VCB in complex with compound 27 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-1-[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
4M7S
 
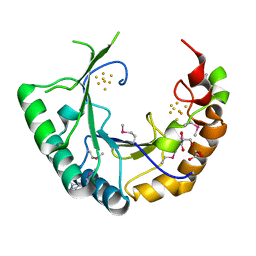 | |
8BDJ
 
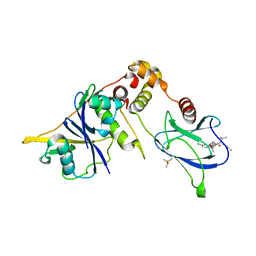 | | VCB in complex with compound 30 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-1-[4-chloranyl-2-(2-methoxyethoxy)phenyl]ethyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
3WQP
 
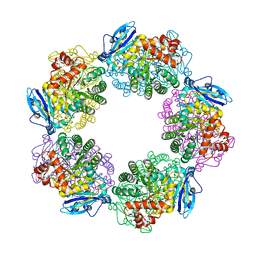 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
4IGM
 
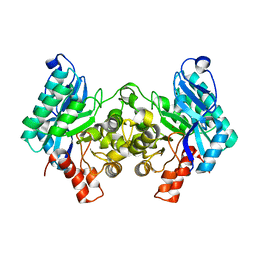 | | 2.39 Angstrom X-ray Crystal structure of human ACMSD | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | 2.39 Angstrom X-ray Crystal structure of human ACMSD
To be Published
|
|
4IJU
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with (1S,4S)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol | | Descriptor: | (1s,4s)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4MDN
 
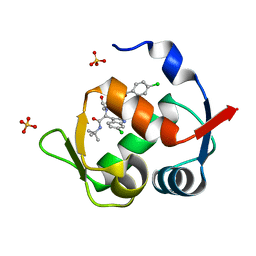 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
4MDT
 
 | | Structure of LpxC bound to the reaction product UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine | | Descriptor: | PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Clayton, G.M, Klein, D.J, Rickert, K.W, Patel, S.B, Kornienko, M, Zugay-Murphy, J, Reid, J.C, Tummala, S, Sharma, S, Singh, S.B, Miesel, L, Lumb, K.J, Soisson, S.M. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer.
J.Biol.Chem., 288, 2013
|
|
4LPL
 
 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
