3F7Q
 
 | |
3IK0
 
 | | Lactobacillus casei Thymidylate Synthase in Ternary Complex with dUMP and the Phtalimidic Derivative 7C1 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzenecarboximidamide, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
5C0V
 
 | | Structure of the LARP1-unique domain DM15 | | Descriptor: | La-related protein 1, SULFATE ION | | Authors: | Lahr, R.M, Berman, A.J. | | Deposit date: | 2015-06-12 | | Release date: | 2015-08-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Nucleic Acids Res., 43, 2015
|
|
5CAJ
 
 | | Crystal structure of E. coli YaaA, a member of the DUF328/UPF0246 family | | Descriptor: | BENZAMIDINE, CHLORIDE ION, UPF0246 protein YaaA | | Authors: | Prahlad, J, Lin, J, Wilson, M.A. | | Deposit date: | 2015-06-29 | | Release date: | 2016-02-24 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The DUF328 family member YaaA is a DNA-binding protein with a novel fold.
J.Biol.Chem., 295, 2020
|
|
1QV6
 
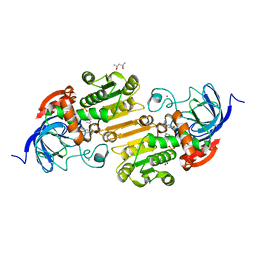 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,4-DIFLUOROBENZYL ALCOHOL | | Descriptor: | (2,4-DIFLUOROPHENYL)METHANOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alcohol dehydrogenase E chain, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-26 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
3AE0
 
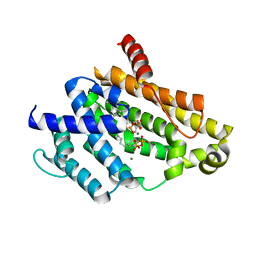 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with geranylgeranyl thiopyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4J2B
 
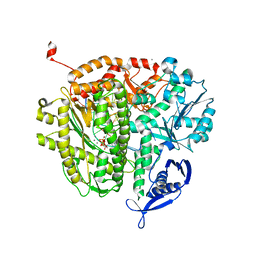 | | RB69 DNA Polymerase L415G Ternary Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*T)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
1FYB
 
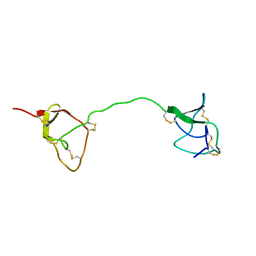 | | SOLUTION STRUCTURE OF C1-T1, A TWO-DOMAIN PROTEINASE INHIBITOR DERIVED FROM THE CIRCULAR PRECURSOR PROTEIN NA-PROPI FROM NICOTIANA ALATA | | Descriptor: | PROTEINASE INHIBITOR | | Authors: | Craik, D.J, Schirra, H.J, Scanlon, M.J, Anderson, M.A. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata.
J.Mol.Biol., 306, 2001
|
|
5K2F
 
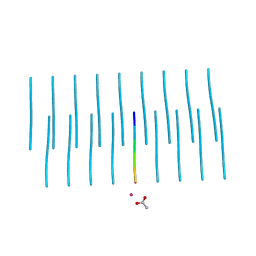 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3A4C
 
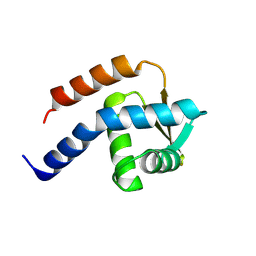 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
1C2R
 
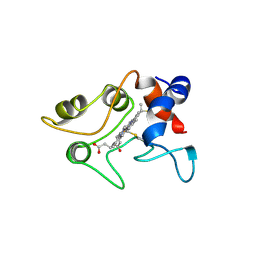 | | MOLECULAR STRUCTURE OF CYTOCHROME C2 ISOLATED FROM RHODOBACTER CAPSULATUS DETERMINED AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Benning, M.M, Wesenberg, G, Caffrey, M.S, Bartsch, R.G, Meyer, T.E, Cusanovich, M.A, Rayment, I, Holden, H.M. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular structure of cytochrome c2 isolated from Rhodobacter capsulatus determined at 2.5 A resolution.
J.Mol.Biol., 220, 1991
|
|
1G9U
 
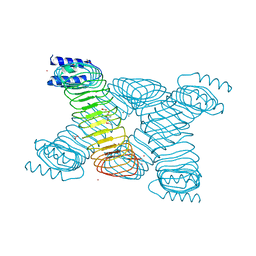 | | CRYSTAL STRUCTURE OF YOPM-LEUCINE RICH EFFECTOR PROTEIN FROM YERSINIA PESTIS | | Descriptor: | ACETATE ION, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Evdokimov, A.G, Anderson, D.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-11-28 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit.
J.Mol.Biol., 312, 2001
|
|
2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
3AA0
 
 | | Crystal structure of Actin Capping Protein in complex with the Cp-binding motif derived from CARMIL | | Descriptor: | 21mer peptide from Leucine-rich repeat-containing protein 16A, CARBONATE ION, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
3A25
 
 | | Crystal structure of P. horikoshii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5UW3
 
 | | PCY1 in Complex with Follower Peptide | | Descriptor: | CACODYLATE ION, Peptide cyclase 1, Presegetalin A1 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UZW
 
 | |
3OIP
 
 | | Crystal structure of Yeast telomere protein Cdc13 OB1 | | Descriptor: | Cell division control protein 13 | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2NAD
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | AZIDE ION, NAD-DEPENDENT FORMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
1CLL
 
 | |
4J2A
 
 | | RB69 DNA Polymerase L415A Ternary Complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
1YFM
 
 | |
3SGA
 
 | |
1QI6
 
 | | SECOND APO FORM OF AN NADP DEPENDENT ALDEHYDE DEHYDROGENASE WITH GLU250 SITUATED 3.7 A FROM CYS284 | | Descriptor: | PROTEIN (NADP DEPENDENT NONPHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1999-06-02 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 300, 2000
|
|
