3NZQ
 
 | | Crystal Structure of Biosynthetic arginine decarboxylase ADC (SpeA) from Escherichia coli, Northeast Structural Genomics Consortium Target ER600 | | Descriptor: | Biosynthetic arginine decarboxylase, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1AWO
 
 | |
1C0P
 
 | | D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES | | Descriptor: | D-ALANINE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Umhau, S, Pollegioni, L, Molla, G, Diederichs, K, Welte, W, Pilone, S.M, Ghisla, S. | | Deposit date: | 1999-07-19 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1A14
 
 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
1D6U
 
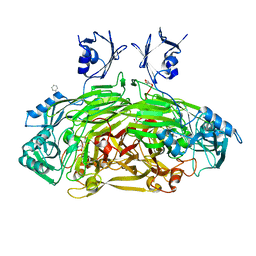 | | CRYSTAL STRUCTURE OF E. COLI AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1D6Z
 
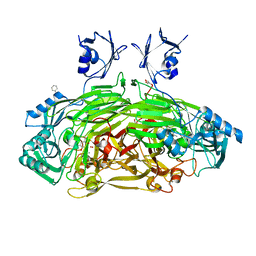 | | CRYSTAL STRUCTURE OF THE AEROBICALLY FREEZE TRAPPED RATE-DETERMINING CATALYTIC INTERMEDIATE OF E. COLI COPPER-CONTAINING AMINE OXIDASE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1D6Y
 
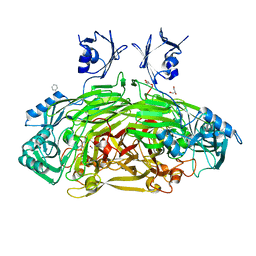 | | CRYSTAL STRUCTURE OF E. COLI COPPER-CONTAINING AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE AND COMPLEXED WITH NITRIC OXIDE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1E2L
 
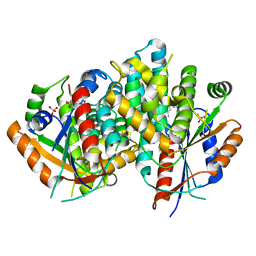 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1E2M
 
 | | HPT + HMTT | | Descriptor: | 6-HYDROXYPROPYLTHYMINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
1CSB
 
 | |
1E2K
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1E2N
 
 | | HPT + HMTT | | Descriptor: | 6-{[4-(HYDROXYMETHYL)-5-METHYL-2,6-DIOXOHEXAHYDROPYRIMIDIN-5-YL]METHYL}-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
1E2P
 
 | | Thymidine kinase, DHBT | | Descriptor: | 6-[3-HYDROXY-2-(HYDROXYMETHYL)PROPYL]-5-METHYL-2,4(1H,3H)-PYRIMIDINEDIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Schulz, G.E, Kessler, U. | | Deposit date: | 2000-05-23 | | Release date: | 2001-04-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
2LV8
 
 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | Descriptor: | De novo designed rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2V87
 
 | |
2V83
 
 | |
3IGN
 
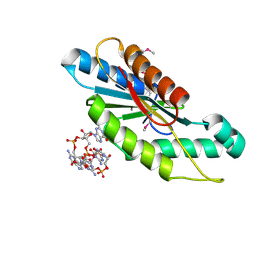 | | Crystal Structure of the GGDEF domain from Marinobacter aquaeolei diguanylate cyclase complexed with c-di-GMP - Northeast Structural Genomics Consortium Target MqR89a | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate cyclase | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product.
J.Struct.Funct.Genom., 13, 2012
|
|
2X83
 
 | |
2IT8
 
 | |
8DUL
 
 | | Cryo-EM Structure of Antibody SKT05 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab SKT05 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DUN
 
 | | Cryo-EM Structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody SKW11 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DWO
 
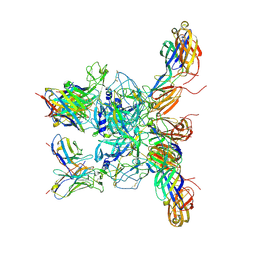 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
2XOT
 
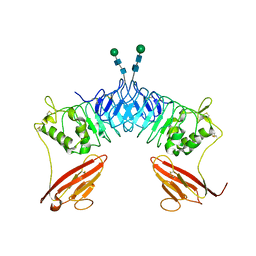 | | Crystal structure of neuronal leucine rich repeat protein AMIGO-1 | | Descriptor: | Amphoterin-induced protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kajander, T, Kuja-Panula, J, Rauvala, H, Goldman, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Role of Glycans and Dimerisation in Folding of Neuronal Leucine-Rich Repeat Protein Amigo-1
J.Mol.Biol., 413, 2011
|
|
2V88
 
 | |
8EEU
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05 | | Descriptor: | Coat protein, Fab SKT05 heavy chain, Fab SKT05 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
