7KSG
 
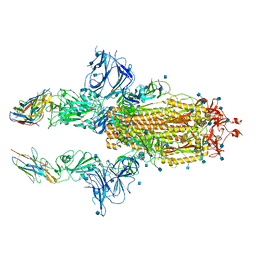 | | SARS-CoV-2 spike in complex with nanobodies E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
6CAW
 
 | |
7E1Z
 
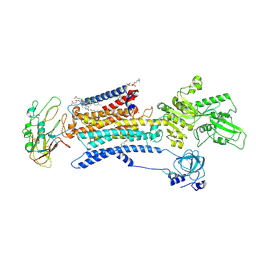 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
4MMU
 
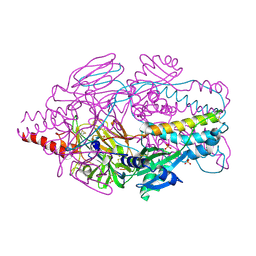 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMT
 
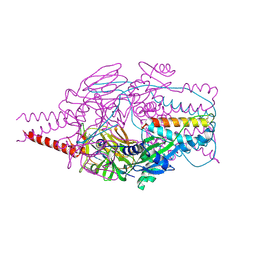 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
6ML8
 
 | |
1S9N
 
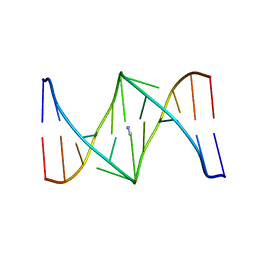 | | Solution structure of the nitrous acid (G)-(G) cross-linked DNA dodecamer duplex GCATCC(G)GATGC | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*CP*GP*GP*AP*TP*GP*C)-3' | | Authors: | Edfeldt, N.B.F, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link
Nucleic Acids Res., 32, 2004
|
|
1IEX
 
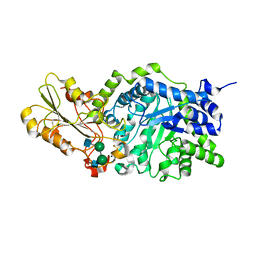 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Driguez, H, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1RER
 
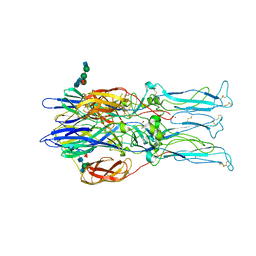 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
1J8V
 
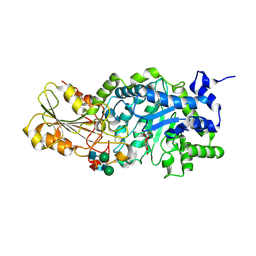 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Fairweather, J.K, Driguez, H, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad substrate specificity in higher plant beta-D-glucan glucohydrolases.
Plant Cell, 14, 2002
|
|
1RHY
 
 | | Crystal structure of Imidazole Glycerol Phosphate Dehydratase | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, GLYCEROL, ... | | Authors: | Sinha, S.C, Chaudhuri, B.N, Burgner, J.W, Yakovleva, G, Davisson, V.J, Smith, J.L. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold
J.Biol.Chem., 279, 2004
|
|
1IHA
 
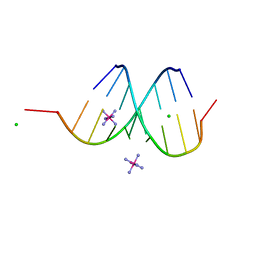 | | Structure of the Hybrid RNA/DNA R-GCUUCGGC-D[BR]U in Presence of RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(BRU))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W.B, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4C55
 
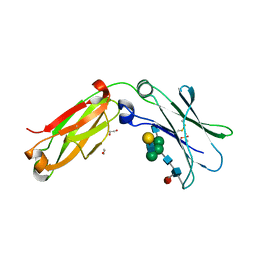 | | Crystal structure of serum-derived human IgG4 Fc | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, IG GAMMA-4 CHAIN C REGION, ... | | Authors: | Davies, A.M, Rispens, T, Ooijevaar-deHeer, P, Gould, H.J, Jefferis, R, Aalberse, R.C, Sutton, B.J. | | Deposit date: | 2013-09-10 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Determinants of Unique Properties of Human Igg4-Fc
J.Mol.Biol., 426, 2014
|
|
1RPQ
 
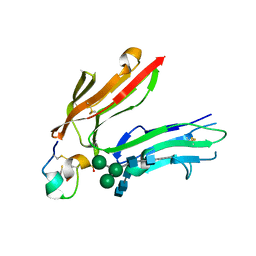 | | High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stamos, J, Eigenbrot, C, Nakamura, G.R, Reynolds, M.E, Yin, J.P, Lowman, H.B, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Convergent Recognition of the IgE Binding Site on the High-Affinity IgE Receptor.
Structure, 12, 2004
|
|
1IGT
 
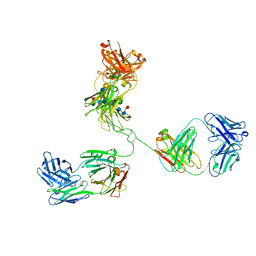 | | STRUCTURE OF IMMUNOGLOBULIN | | Descriptor: | IGG2A INTACT ANTIBODY - MAB231, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Harris, L.J, Larson, S.B, Hasel, K.W, McPherson, A. | | Deposit date: | 1996-10-25 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Refined structure of an intact IgG2a monoclonal antibody.
Biochemistry, 36, 1997
|
|
1IEW
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
4BSV
 
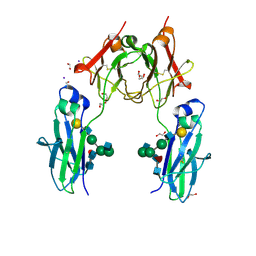 | | Heterodimeric Fc Antibody Azymetric Variant 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, HETERODIMERIC FC ANTIBODY AZYMETRIC VARIANT 2, ... | | Authors: | Suits, M.D.L, Spreter, T, Cabrera, E.E, Dixit, S.B, Lario, P.I, Poon, D.K.Y, D'Angelo, I.E.P, Boulanger, M.J. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Improving Biophysical Properties of a Bispecific Antibody Scaffold to Aid Developability: Quality by Molecular Design.
Mabs, 5, 2013
|
|
1S9O
 
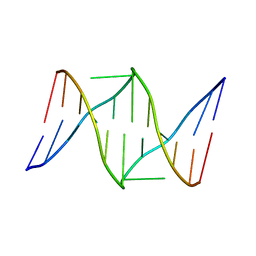 | | Solution structure of the nitrous acid induced DNA interstrand cross-linked dodecamer duplex CGCTAC(G)TAGCG with the cross-linked guanines denoted (G) | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*G)-3' | | Authors: | Edfeldt, N.B, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link.
Nucleic Acids Res., 32, 2004
|
|
2RTP
 
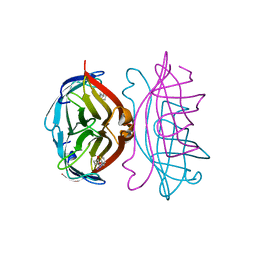 | |
2RTB
 
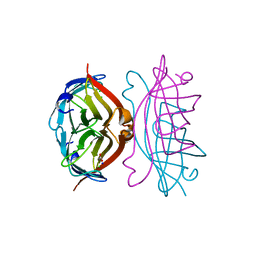 | | APOSTREPTAVIDIN, PH 3.32, SPACE GROUP I222 | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTH
 
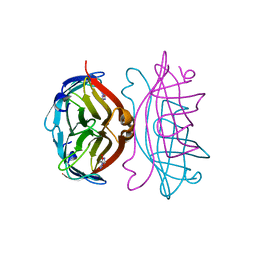 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I222 | | Descriptor: | ACETATE ION, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTE
 
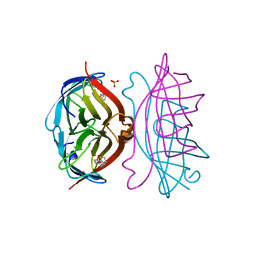 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 1.90, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTF
 
 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 2.00, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
5IQU
 
 | | WelO5 G166D variant bound to Fe(II), 2-oxoglutarate, and 12-epifischerindole U | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mitchell, A.J, Maggiolo, A.O, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
