2FEU
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
2FT7
 
 | |
2FTA
 
 | |
2FSF
 
 | |
2FNA
 
 | |
2FYE
 
 | | Mutant Human Cathepsin S with irreversible inhibitor CRA-14013 | | Descriptor: | N-[(1R)-1-[({[(5S)-3,5-DIMETHYL-2,5-DIHYDROISOXAZOL-4-YL]METHYL}SULFONYL)METHYL]-2-OXO-2-({(1S)-3-PHENYL-1-[2-(PHENYLSULFONYL)ETHYL]PROPYL}AMINO)ETHYL]MORPHOLINE-4-CARBOXAMIDE, cathepsin S preproprotein | | Authors: | Somoza, J.R. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Human Cathepsin S with irreversible inhibitor CRA-14013
To be Published
|
|
2FR4
 
 | | Structure of Fab DNA-1 complexed with a stem-loop DNA ligand | | Descriptor: | 5'-D(*CP*TP*GP*CP*CP*TP*TP*CP*AP*G)-3', TETRAETHYLENE GLYCOL, antibody heavy chain FAB, ... | | Authors: | Tanner, J.J, Ou, Z. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Impact of DNA hairpin folding energetics on antibody-ssDNA association.
J.Mol.Biol., 374, 2007
|
|
2FSI
 
 | |
2FV9
 
 | | Crystal structure of TACE in complex with JMV 390-1 | | Descriptor: | ADAM 17, N-[(2R)-2-BENZYL-4-(HYDROXYAMINO)-4-OXOBUTANOYL]-L-ISOLEUCYL-L-LEUCINE, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2006-01-30 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Stabilization of the autoproteolysis of TNF-alpha converting enzyme (TACE) results in a novel crystal form suitable for structure-based drug design studies.
Protein Eng.Des.Sel., 19, 2006
|
|
2G07
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, phospho-enzyme intermediate analog with Beryllium fluoride | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G40
 
 | |
2FZN
 
 | |
2G06
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, with bound magnesium(II) | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G0T
 
 | |
2G36
 
 | |
2G80
 
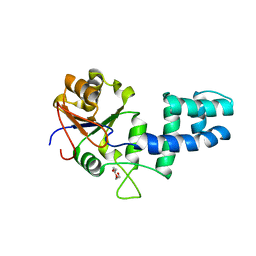 | |
2GA4
 
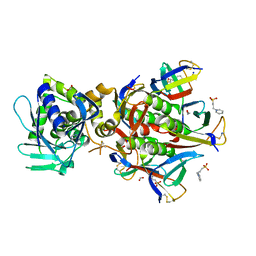 | | Stx2 with adenine | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ADENINE, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2006-03-07 | | Release date: | 2006-07-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of adenine to Stx2, the protein toxin from Escherichia coli O157:H7.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2FQE
 
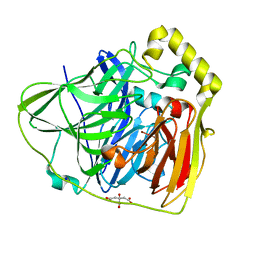 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FS6
 
 | |
2FSY
 
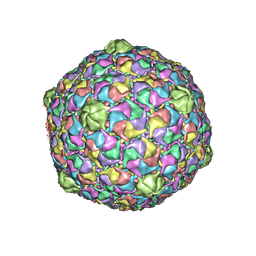 | | Bacteriophage HK97 Pepsin-treated Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2FVG
 
 | |
2FY6
 
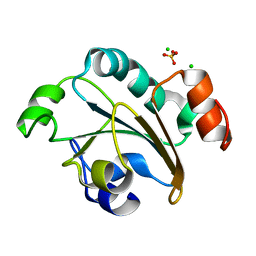 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
2ETS
 
 | |
2F1N
 
 | |
2F22
 
 | |
