6N19
 
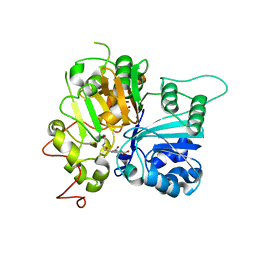 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-carboxybutanoyl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
1XXD
 
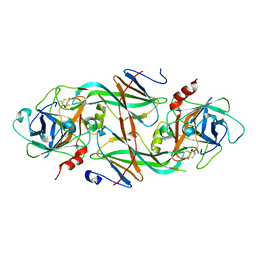 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | Descriptor: | Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
2GHE
 
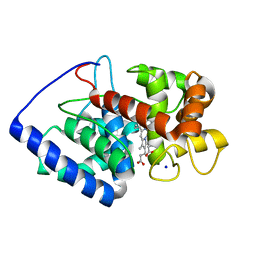 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, cytosolic ascorbate peroxidase 1 | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
1UKL
 
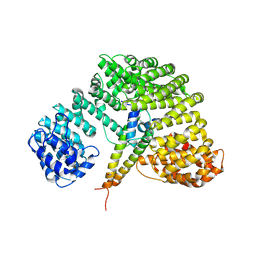 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
1XX9
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with EcotinM84R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
4OO1
 
 | |
1SN5
 
 | | Crystal Structure of Sea Bream Transthyretin in complex with Triiodothyronine at 1.90A Resolution | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1D8F
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY-1-(4-METHOXYPHENYL)SULFONYL-4-BENZYLOXYCARBONYL-PIPERAZINE-2-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, De, B, Pikul, S, Almstead, N.G, Natchus, M.G, Anastasio, M.V, McPhail, S.J, Snider, C.E, Taiwo, Y.O, Chen, L.Y. | | Deposit date: | 1999-10-22 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of piperazine-based matrix metalloproteinase inhibitors.
J.Med.Chem., 43, 2000
|
|
1SOJ
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B IN COMPLEX WITH IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, cGMP-inhibited 3',5'-cyclic phosphodiesterase B | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1SHH
 
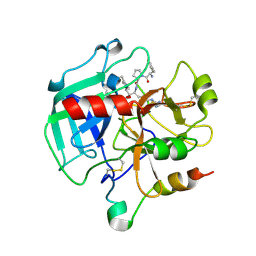 | | Slow form of Thrombin Bound with PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-25 | | Release date: | 2004-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
1SL6
 
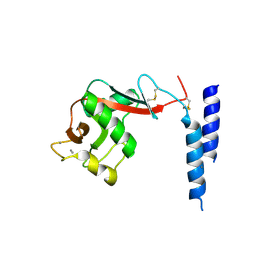 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SN2
 
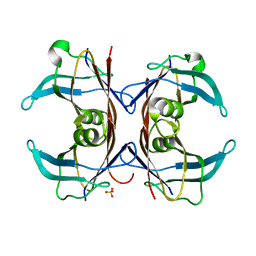 | | Crystal Structure of Sea Bream Transthyretin at 1.90A Resolution | | Descriptor: | SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
2DTZ
 
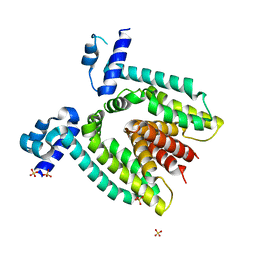 | |
1CW2
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-25 | | Release date: | 1999-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1XJA
 
 | |
1R18
 
 | | Drosophila protein isoaspartyl methyltransferase with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate(D-aspartate)-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bennett, E.J, Bjerregaard, J, Knapp, J.E, Chavous, D.A, Friedman, A.M, Royer Jr, W.E, O'Connor, C.M. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis.
Biochemistry, 42, 2003
|
|
1SHQ
 
 | | Crystal structure of shrimp alkaline phosphatase with magnesium in M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SHN
 
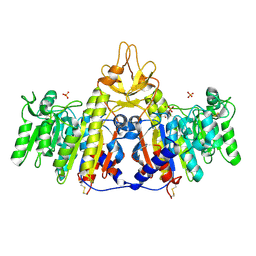 | | Crystal structure of shrimp alkaline phosphatase with phosphate bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RZU
 
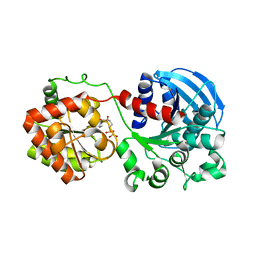 | | Crystal structure of the glycogen synthase from A. tumefaciens in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase 1 | | Authors: | Buschiazzo, A, Guerin, M.E, Ugalde, J.E, Ugalde, R.A, Shepard, W, Alzari, P.M. | | Deposit date: | 2003-12-29 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation.
Embo J., 23, 2004
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
2QJM
 
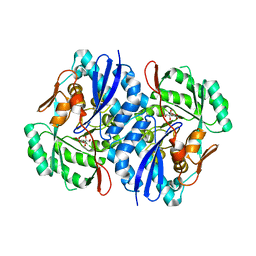 | | Crystal structure of the K271E mutant of Mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and D-mannonate | | Descriptor: | D-MANNONIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2GJ6
 
 | |
4FZV
 
 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
2QJJ
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
