8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5KLZ
 
 | |
8QG8
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]guanidine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG4
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-naphthalen-1-yl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGC
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-phenyl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-pent-4-ynyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGI
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-(phenylmethyl)urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGO
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
5NR9
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | Descriptor: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5KM1
 
 | |
7NOY
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis in complex with substrate 1-hydroxyquinolin-4(1H)-one | | Descriptor: | 1-oxidanylquinolin-4-one, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
6FMV
 
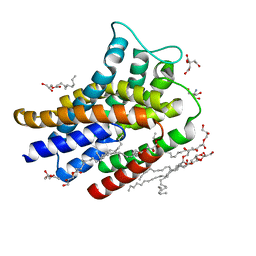 | | IMISX-EP of Hg-BacA soaking SIRAS | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
8U6H
 
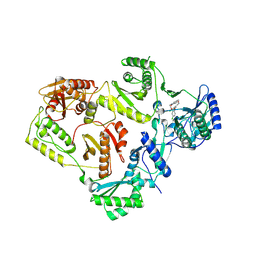 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8XM4
 
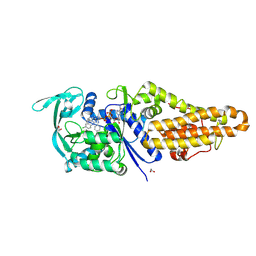 | | Methionyl-tRNA synthetase from Staphylococcus aureus in complex with chlorhexidine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lu, F, Xia, K, Yi, J, Chen, B, Luo, Z, Xu, J, Gu, Q, Zhou, H. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and structural characterization of chlorhexidine as an ATP-assisted inhibitor against type 1 methionyl-tRNA synthetase from Gram-positive bacteria.
Eur.J.Med.Chem., 268, 2024
|
|
7UFY
 
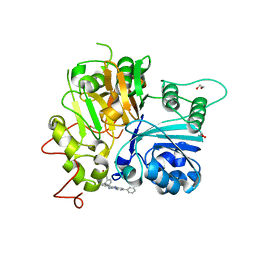 | | Crystal structure of TDP1 complexed with compound XZ766 | | Descriptor: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, [(4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)methyl]phosphonic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
6HX1
 
 | | IRE1 ALPHA IN COMPLEX WITH imidazo[1,2-b]pyridazin-8-amine compound 2 | | Descriptor: | 6-chloranyl-~{N}-(cyclopropylmethyl)-3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Augustin, M.A, Krapp, S, Bayliss, R, Collins, I. | | Deposit date: | 2018-10-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
5MW8
 
 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
6RZF
 
 | | Galectin-3C in complex with ortho-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.016 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
5OHH
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, ACETATE ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of human carbonic anhydrase isozyme XIII with 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To be published
|
|
6FMW
 
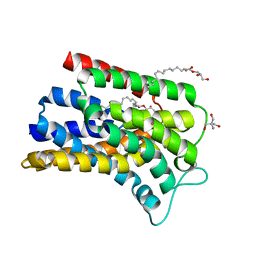 | | IMISX-EP of Hg-BacA cocrystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
8D0S
 
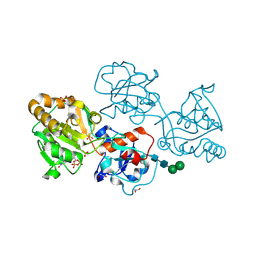 | | Human FUT9 bound to GDP and LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8AYF
 
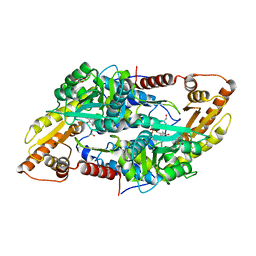 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
8D0X
 
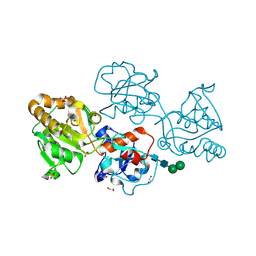 | | Human FUT9 bound to LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0R
 
 | | Human FUT9 bound to GDP and H-Type 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
